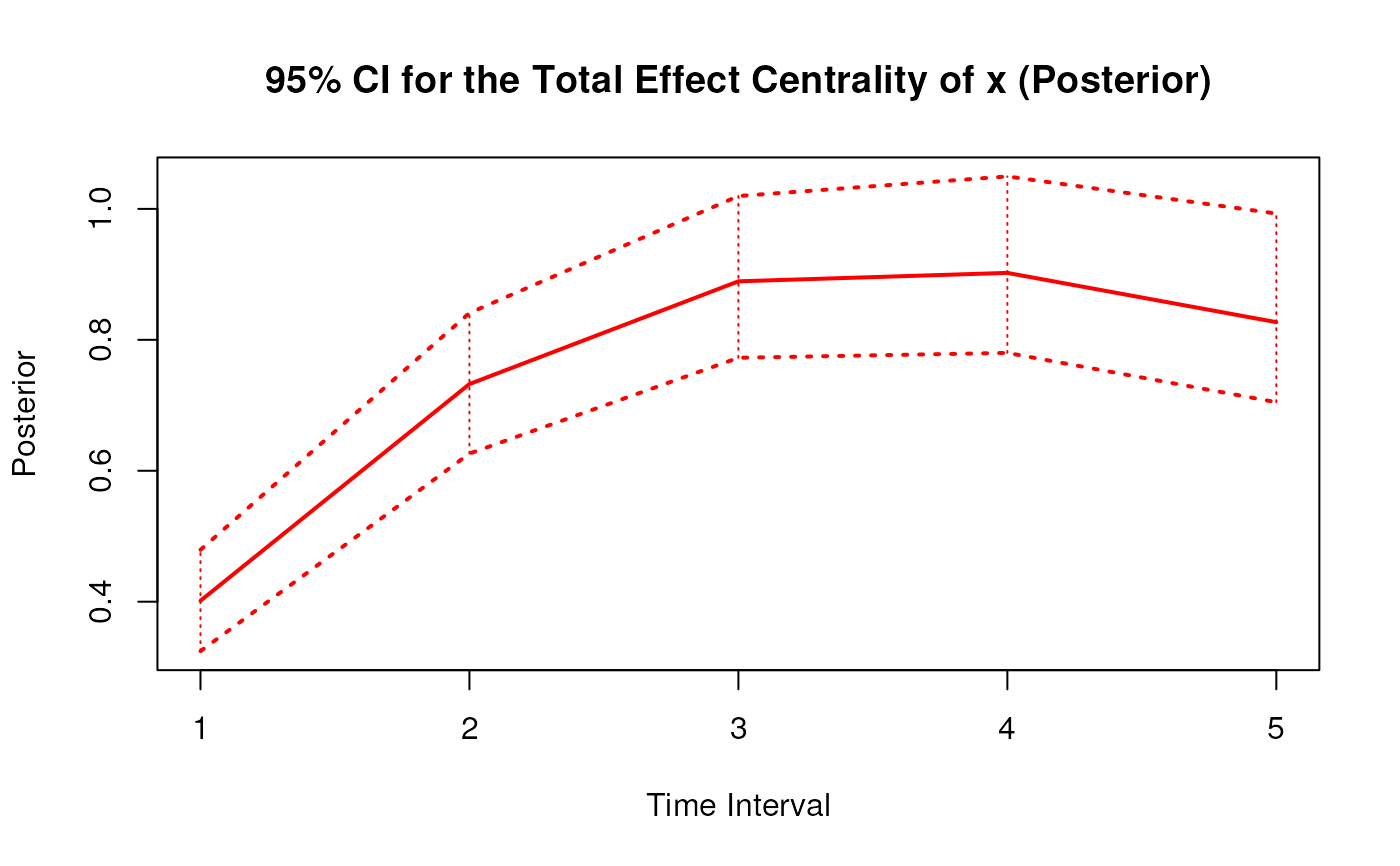
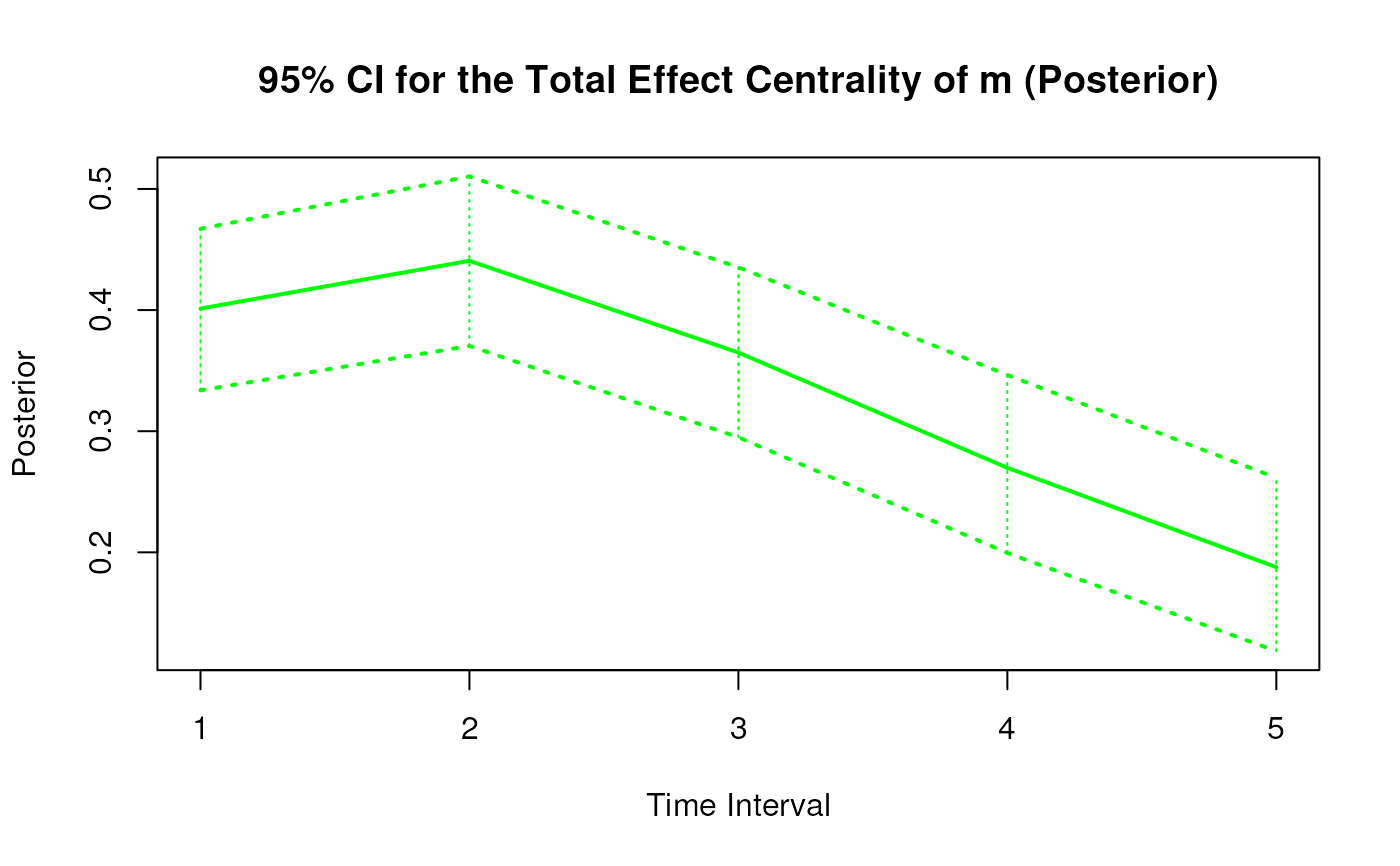
Posterior Distribution of the Total Effect Centrality Over a Specific Time Interval or a Range of Time Intervals
Source:R/cTMed-posterior-total-central.R
PosteriorTotalCentral.RdThis function generates a posterior distribution of the total effect centrality over a specific time interval \(\Delta t\) or a range of time intervals using the posterior distribution of the first-order stochastic differential equation model drift matrix \(\boldsymbol{\Phi}\).
Arguments
- phi
List of numeric matrices. Each element of the list is a sample from the posterior distribution of the drift matrix (\(\boldsymbol{\Phi}\)). Each matrix should have row and column names pertaining to the variables in the system.
- delta_t
Numeric. Time interval (\(\Delta t\)).
- ncores
Positive integer. Number of cores to use. If
ncores = NULL, use a single core. Consider using multiple cores when number of replicationsRis a large value.- tol
Numeric. Smallest possible time interval to allow.
Value
Returns an object
of class ctmedmc which is a list with the following elements:
- call
Function call.
- args
Function arguments.
- fun
Function used ("PosteriorTotalCentral").
- output
A list the length of which is equal to the length of
delta_t.
Each element in the output list has the following elements:
- est
Mean of the posterior distribution of the total, direct, and indirect effects.
- thetahatstar
Posterior distribution of the total, direct, and indirect effects.
Details
See TotalCentral() for more details.
References
Bollen, K. A. (1987). Total, direct, and indirect effects in structural equation models. Sociological Methodology, 17, 37. doi:10.2307/271028
Deboeck, P. R., & Preacher, K. J. (2015). No need to be discrete: A method for continuous time mediation analysis. Structural Equation Modeling: A Multidisciplinary Journal, 23 (1), 61-75. doi:10.1080/10705511.2014.973960
Pesigan, I. J. A., Russell, M. A., & Chow, S.-M. (2025). Inferences and effect sizes for direct, indirect, and total effects in continuous-time mediation models. Psychological Methods. doi:10.1037/met0000779
Ryan, O., & Hamaker, E. L. (2021). Time to intervene: A continuous-time approach to network analysis and centrality. Psychometrika, 87 (1), 214-252. doi:10.1007/s11336-021-09767-0
See also
Other Continuous-Time Mediation Functions:
BootBeta(),
BootBetaStd(),
BootIndirectCentral(),
BootMed(),
BootMedStd(),
BootTotalCentral(),
DeltaBeta(),
DeltaBetaStd(),
DeltaIndirectCentral(),
DeltaMed(),
DeltaMedStd(),
DeltaTotalCentral(),
Direct(),
DirectStd(),
Indirect(),
IndirectCentral(),
IndirectStd(),
MCBeta(),
MCBetaStd(),
MCIndirectCentral(),
MCMed(),
MCMedStd(),
MCPhi(),
MCPhiSigma(),
MCTotalCentral(),
Med(),
MedStd(),
PosteriorBeta(),
PosteriorIndirectCentral(),
PosteriorMed(),
Total(),
TotalCentral(),
TotalStd(),
Trajectory()
Examples
phi <- matrix(
data = c(
-0.357, 0.771, -0.450,
0.0, -0.511, 0.729,
0, 0, -0.693
),
nrow = 3
)
colnames(phi) <- rownames(phi) <- c("x", "m", "y")
vcov_phi_vec <- matrix(
data = c(
0.00843, 0.00040, -0.00151,
-0.00600, -0.00033, 0.00110,
0.00324, 0.00020, -0.00061,
0.00040, 0.00374, 0.00016,
-0.00022, -0.00273, -0.00016,
0.00009, 0.00150, 0.00012,
-0.00151, 0.00016, 0.00389,
0.00103, -0.00007, -0.00283,
-0.00050, 0.00000, 0.00156,
-0.00600, -0.00022, 0.00103,
0.00644, 0.00031, -0.00119,
-0.00374, -0.00021, 0.00070,
-0.00033, -0.00273, -0.00007,
0.00031, 0.00287, 0.00013,
-0.00014, -0.00170, -0.00012,
0.00110, -0.00016, -0.00283,
-0.00119, 0.00013, 0.00297,
0.00063, -0.00004, -0.00177,
0.00324, 0.00009, -0.00050,
-0.00374, -0.00014, 0.00063,
0.00495, 0.00024, -0.00093,
0.00020, 0.00150, 0.00000,
-0.00021, -0.00170, -0.00004,
0.00024, 0.00214, 0.00012,
-0.00061, 0.00012, 0.00156,
0.00070, -0.00012, -0.00177,
-0.00093, 0.00012, 0.00223
),
nrow = 9
)
phi <- MCPhi(
phi = phi,
vcov_phi_vec = vcov_phi_vec,
R = 1000L
)$output
# Specific time interval ----------------------------------------------------
PosteriorTotalCentral(
phi = phi,
delta_t = 1
)
#> Call:
#> PosteriorTotalCentral(phi = phi, delta_t = 1)
#>
#> Total Effect Centrality
#>
#> variable interval est se R 2.5% 97.5%
#> 1 x 1 0.3991 0.0485 1000 0.3039 0.4915
#> 2 m 1 0.3987 0.0416 1000 0.3158 0.4763
#> 3 y 1 0.0017 0.0651 1000 -0.1237 0.1280
# Range of time intervals ---------------------------------------------------
posterior <- PosteriorTotalCentral(
phi = phi,
delta_t = 1:5
)
# Methods -------------------------------------------------------------------
# PosteriorTotalCentral has a number of methods including
# print, summary, confint, and plot
print(posterior)
#> Call:
#> PosteriorTotalCentral(phi = phi, delta_t = 1:5)
#>
#> Total Effect Centrality
#>
#> variable interval est se R 2.5% 97.5%
#> 1 x 1 0.3991 0.0485 1000 0.3039 0.4915
#> 2 m 1 0.3987 0.0416 1000 0.3158 0.4763
#> 3 y 1 0.0017 0.0651 1000 -0.1237 0.1280
#> 4 x 2 0.7324 0.0695 1000 0.5981 0.8725
#> 5 m 2 0.4386 0.0536 1000 0.3283 0.5424
#> 6 y 2 0.0020 0.0952 1000 -0.1858 0.1888
#> 7 x 3 0.8935 0.0880 1000 0.7298 1.0839
#> 8 m 3 0.3631 0.0612 1000 0.2403 0.4838
#> 9 y 3 0.0015 0.1027 1000 -0.2063 0.2047
#> 10 x 4 0.9099 0.1027 1000 0.7253 1.1318
#> 11 m 4 0.2680 0.0662 1000 0.1298 0.3954
#> 12 y 4 0.0012 0.0971 1000 -0.1927 0.1928
#> 13 x 5 0.8368 0.1122 1000 0.6337 1.0768
#> 14 m 5 0.1862 0.0682 1000 0.0471 0.3200
#> 15 y 5 0.0012 0.0851 1000 -0.1688 0.1678
summary(posterior)
#> Call:
#> PosteriorTotalCentral(phi = phi, delta_t = 1:5)
#>
#> Total Effect Centrality
#>
#> variable interval est se R 2.5% 97.5%
#> 1 x 1 0.3991 0.0485 1000 0.3039 0.4915
#> 2 m 1 0.3987 0.0416 1000 0.3158 0.4763
#> 3 y 1 0.0017 0.0651 1000 -0.1237 0.1280
#> 4 x 2 0.7324 0.0695 1000 0.5981 0.8725
#> 5 m 2 0.4386 0.0536 1000 0.3283 0.5424
#> 6 y 2 0.0020 0.0952 1000 -0.1858 0.1888
#> 7 x 3 0.8935 0.0880 1000 0.7298 1.0839
#> 8 m 3 0.3631 0.0612 1000 0.2403 0.4838
#> 9 y 3 0.0015 0.1027 1000 -0.2063 0.2047
#> 10 x 4 0.9099 0.1027 1000 0.7253 1.1318
#> 11 m 4 0.2680 0.0662 1000 0.1298 0.3954
#> 12 y 4 0.0012 0.0971 1000 -0.1927 0.1928
#> 13 x 5 0.8368 0.1122 1000 0.6337 1.0768
#> 14 m 5 0.1862 0.0682 1000 0.0471 0.3200
#> 15 y 5 0.0012 0.0851 1000 -0.1688 0.1678
confint(posterior, level = 0.95)
#> variable interval 2.5 % 97.5 %
#> 1 x 1 0.30389727 0.4914777
#> 2 m 1 0.31584766 0.4762828
#> 3 y 1 -0.12372017 0.1279576
#> 4 x 2 0.59814836 0.8724793
#> 5 m 2 0.32826858 0.5424170
#> 6 y 2 -0.18582907 0.1887818
#> 7 x 3 0.72979419 1.0838910
#> 8 m 3 0.24025621 0.4838154
#> 9 y 3 -0.20628187 0.2047223
#> 10 x 4 0.72530366 1.1318121
#> 11 m 4 0.12984662 0.3954040
#> 12 y 4 -0.19272102 0.1928162
#> 13 x 5 0.63366101 1.0767630
#> 14 m 5 0.04707667 0.3200269
#> 15 y 5 -0.16878260 0.1677510
plot(posterior)