The following is a simple benchmark comparing the computational requirements of the methods to generate confidence intervals for the indirect effect with missing observations. We will use the generated data in the Data Generation article.
In this benchmark, we compare the following methods
- Monte Carlo method using full-information maximum likelihood
(
MCML()), - Monte Carlo method using multiple imputation
(
MCMI()), - full-information maximum likelihood nested within nonparametric
bootstrap (
NBML()), - multiple imputation nested within nonparametric bootstrap
(
NBMI()) - nonparametric bootstrap nested within multiple imputation
(
MINB())
Arguments
| Variables | Values | Notes |
|---|---|---|
| m | 100 | Number of imputations. |
| R | 5000 | Number of Monte Carlo replications. |
| B | 5000 | Number of bootstrap samples. |
| mplus_bin | “/opt/mplusdemo/mpdemo” | Path to Mplus binary. |
NOTE: If you are using
manmcmedmiss-rockerormanmcmedmiss.sifdescribed in the Containers article, setmplus_bin = "mpdemo".
Parameters
| Variable | Value | Notes |
|---|---|---|
| n | 50 | \(n\) |
| tauprime | 0.14142135623731 | \(\tau^{\prime}\) |
| alpha | 0.714074191775111 | \(\alpha\) |
| beta | 0.714074191775111 | \(\beta\) |
Data
Amputation
Generate sample data with missing values using the multivariate amputation approach proposed by Schouten et al. (2018).
data_missing <- AmputeData(
data_complete,
mech = "MAR",
prop = 0.10
)Imputation
Perform multiple imputation following Asparouhov and Muthen (2010)
using Mplus.
data_mi <- ImputeData(
data_missing,
m = m,
mplus_bin = mplus_bin
)Maximum Likelihood
Missing Data
Parameters of the simple mediation model are estimated using full-information maximum likelihood to handle missing data.
fit_ml <- FitModelML(
data_missing,
mplus_bin = mplus_bin
)Multiple Imputation
Parameters of the simple mediation model are estimated using maximum likelihood for each of the imputed data sets. The parameter estimates and their sampling covariance matrix are pooled.
fit_mi <- FitModelMI(
data_mi,
mplus_bin = mplus_bin
)Summary of Benchmark Results
summary(benchmark, unit = "ms")
#> expr min lq mean median uq
#> 1 MCML 18.73027 34.62104 48.37869 46.02422 5.337519e+01
#> 2 MCMI 32.08413 34.56102 68.64550 61.40423 1.054957e+02
#> 3 NBML 1779.85401 1995.39254 2686.87359 2700.63207 3.396238e+03
#> 4 NBMI 626443.36697 787734.54704 907889.17563 818272.44441 1.090418e+06
#> 5 MINB 43613.55303 46511.95203 60975.13838 48293.08648 8.038919e+04
#> max neval cld
#> 1 101.9532 10 a
#> 2 123.1622 10 a
#> 3 3541.1389 10 a
#> 4 1263013.0070 10 b
#> 5 92118.3821 10 aSummary of Benchmark Results Relative to the Fastest Method
summary(benchmark, unit = "relative")
#> expr min lq mean median uq
#> 1 MCML 1.000000 1.000000e+00 1.00000 1.000000 1.000000
#> 2 MCMI 1.712956 9.982665e-01 1.41892 1.334172 1.976494
#> 3 NBML 95.025549 5.763526e+01 55.53837 58.678492 63.629527
#> 4 NBMI 33445.510068 2.275306e+04 18766.30431 17779.168542 20429.300198
#> 5 MINB 2328.506620 1.343459e+03 1260.37190 1049.297126 1506.115228
#> max neval cld
#> 1 1.000000 10 a
#> 2 1.208027 10 a
#> 3 34.732974 10 a
#> 4 12388.160554 10 b
#> 5 903.535673 10 aPlot
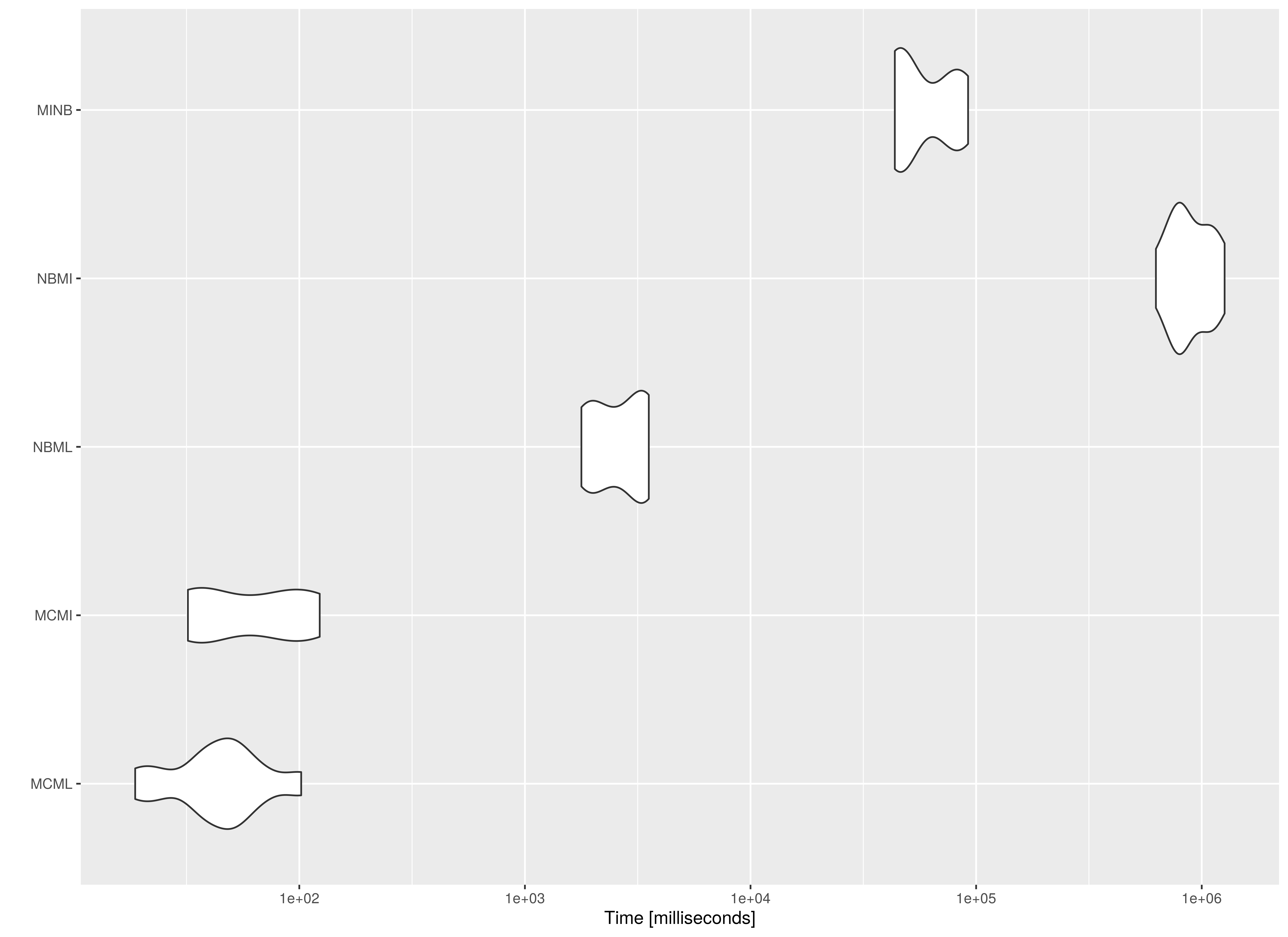
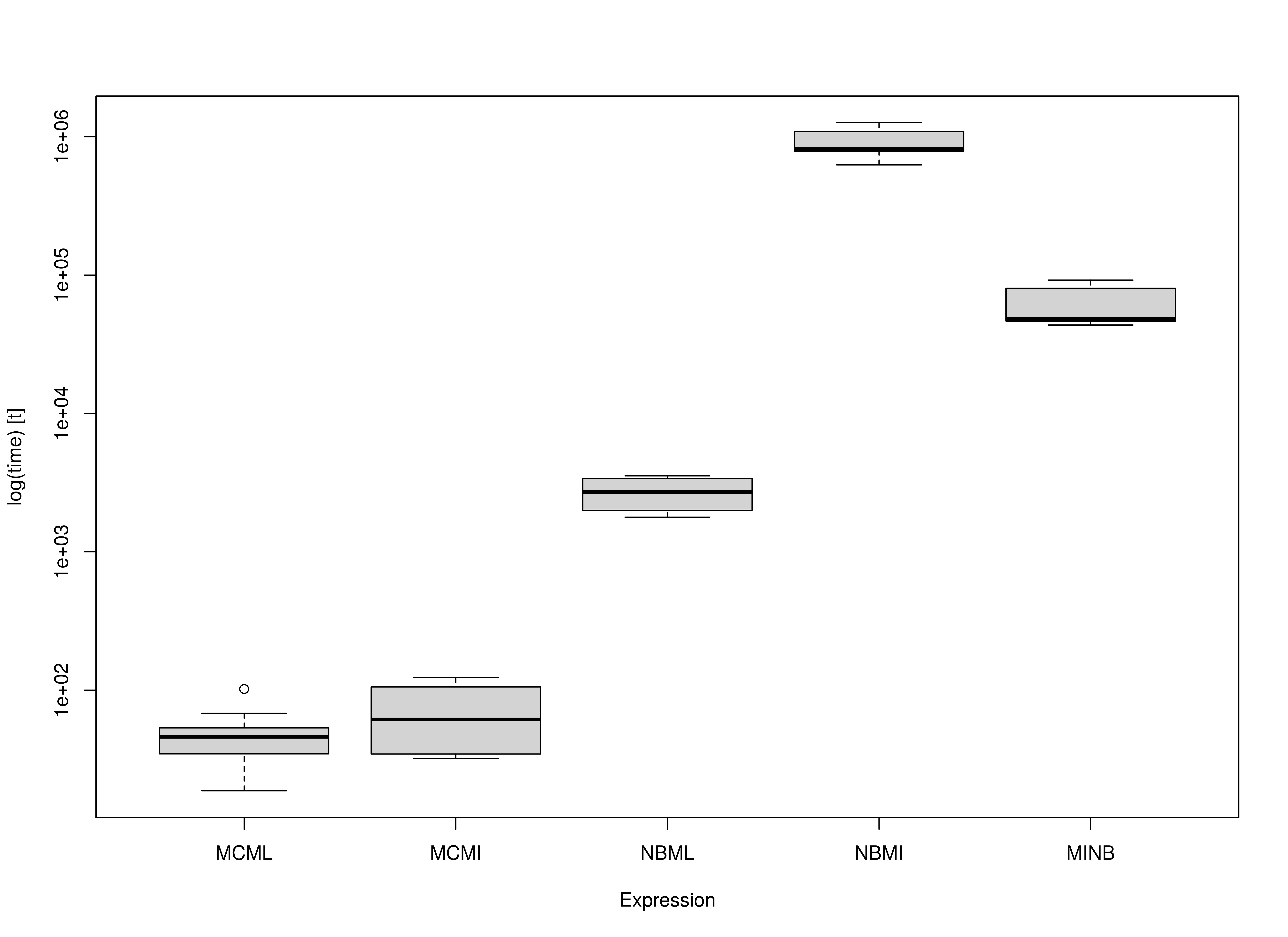
The MC approaches are faster compared to their
NB counterparts. Note the increasing model complexity will
increase the computational cost of NB. However, for
MC, model complexity will not increase the computational
cost of the simulation stage. For example, MI estimates are
more computationally intensive than ML estimates. This
results in a large difference between NBML and the two
NB methods using MI, that is,
NBMI and MINB. Note that MINB is
faster than NBMI as expected but it is still significantly
slower than the MC approaches.
However, if we perform the model fitting step outside the benchmark
calculation, the speed of MCML and MCMI will
be virtually identical. In this implementation, however,
MCMI will be a little bit slower than MCML
because it generates two sets of confidence intervals (vcov
and vcov_tilde) while MCML generates a single
set. Since MC relies on a single estimate of the parameters
and the sampling covariance matrix, it is suited for more complex
models.
NOTE: Note that since
NBonly needs point estimates, a closed form solution of the indirect effect is used inNBMIandMINB. When optimization is used to estimate parameters in the context of structural equation modeling,NBMIandMINBwill be significantly slower.