MCStd Function Use Case 3: R-Squared and Adjusted R-Squared
Ivan Jacob Agaloos Pesigan
2025-10-19
Source:vignettes/mcstd-3-rsqr.Rmd
mcstd-3-rsqr.RmdThe MCStd() function is used to generate Monte Carlo
confidence intervals for
and adjusted
.
Data
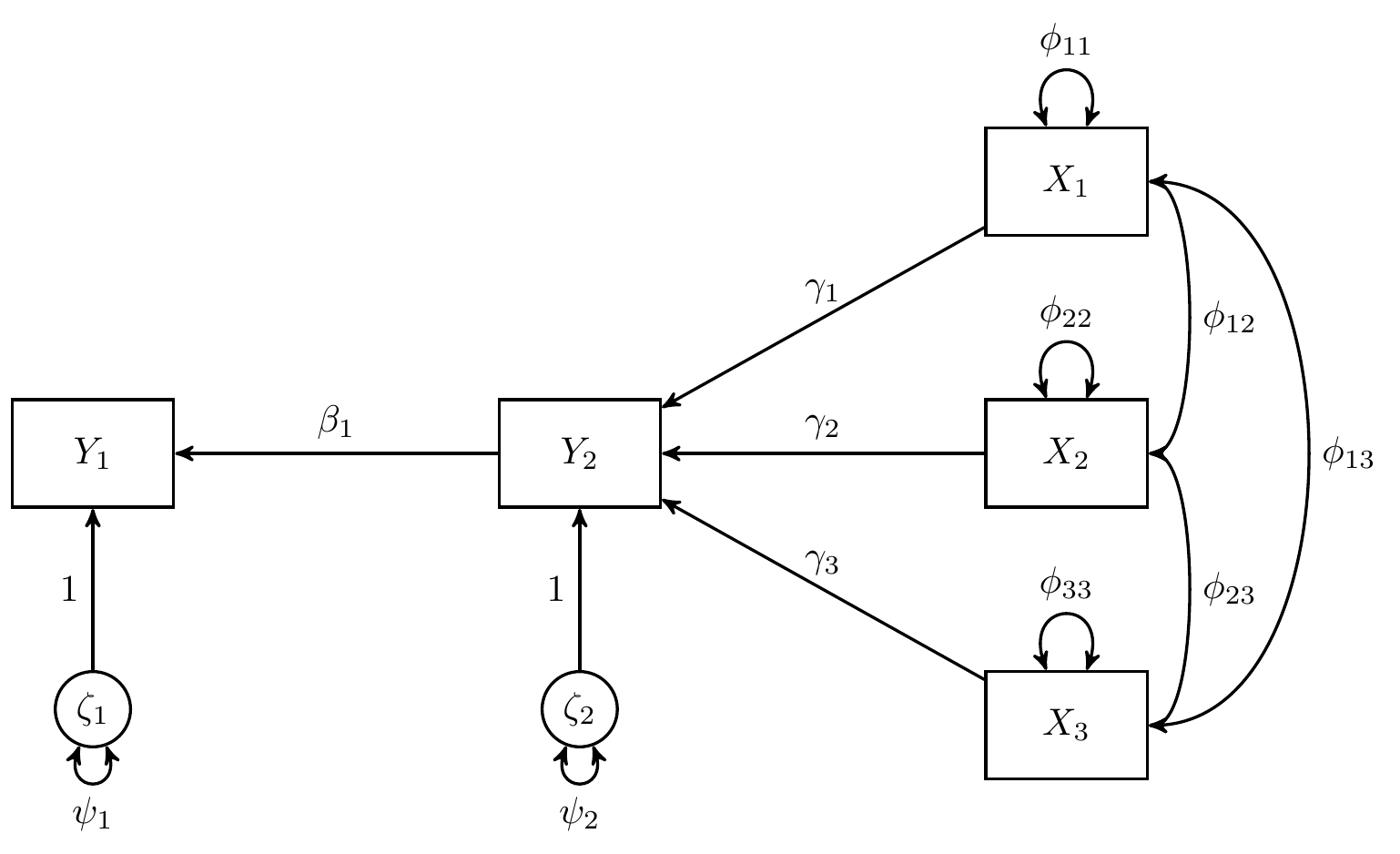
In this example, we use data from Kwan & Chan (2011) where child’s reading ability () is regressed on home educational resources and home educational resources () is regressed on parental occupational status (), parental educational level (), and child’s home possession ()
Note that and are stochastic error terms with expected value of zero and finite variance and , and are intercepts, and , , , and are regression coefficients.
covs
#> Y1 Y2 X1 X2 X3
#> Y1 6088.8281 15.7012 271.1429 49.5848 20.0337
#> Y2 15.7012 0.7084 1.9878 1.0043 0.2993
#> X1 271.1429 1.9878 226.2577 29.9232 4.8812
#> X2 49.5848 1.0043 29.9232 9.0692 1.0312
#> X3 20.0337 0.2993 4.8812 1.0312 0.8371
nobs
#> [1] 200Model Specification
We regress Y1 on Y2 and Y2 on
X1, X2, and X3. We label the
error variances as psi1 and psi2.
and
are defined using the := operator in the
lavaan model syntax using the following equations
where is the standardized error variance, is the sample size, and is the number of regressor variables.
model <- "
Y1 ~ Y2
Y2 ~ X1 + X2 + X3
Y1 ~~ psi1 * Y1
Y2 ~~ psi2 * Y2
rsq1 := 1 - psi1
rsqbar1 := 1 - ((200 - 1) / (200 - 1 + 1)) * (1 - rsq1)
rsq2 := 1 - psi2
rsqbar2 := 1 - ((200 - 1) / (200 - 3 + 1)) * (1 - rsq2)
"Model Fitting
We can now fit the model using the sem() function from
lavaan with mimic = "eqs" to ensure
compatibility with results from Kwan & Chan
(2011).
Note: We recommend setting
fixed.x = FALSEwhen generating standardized estimates and confidence intervals to model the variances and covariances of the exogenous observed variables if they are assumed to be random. Iffixed.x = TRUE, which is the default setting inlavaan,MC()will fix the variances and the covariances of the exogenous observed variables to the sample values.
fit <- sem(
model = model, mimic = "eqs", fixed.x = FALSE,
sample.cov = covs, sample.nobs = nobs
)Standardized Monte Carlo Confidence Intervals
Standardized Monte Carlo Confidence intervals can be generated by
passing the result of the MC() function to the
MCStd() function.
Note: The parameterization of and above should only be interpreted using the output of the
MCStd()function since the input in the functions defined by:=require standardized estimates.
unstd <- MC(fit, R = 20000L, alpha = 0.05)
MCStd(unstd, alpha = 0.05)
#> Standardized Monte Carlo Confidence Intervals
#> est se R 2.5% 97.5%
#> Y1~Y2 0.2391 0.0666 20000 0.1066 0.3659
#> Y2~X1 -0.2449 0.0813 20000 -0.4029 -0.0825
#> Y2~X2 0.4419 0.0793 20000 0.2819 0.5939
#> Y2~X3 0.3101 0.0647 20000 0.1787 0.4330
#> psi1 0.9428 0.0322 20000 0.8661 0.9886
#> psi2 0.7428 0.0530 20000 0.6291 0.8351
#> X1~~X1 1.0000 0.0000 20000 1.0000 1.0000
#> X1~~X2 0.6606 0.0408 20000 0.5743 0.7335
#> X1~~X3 0.3547 0.0629 20000 0.2261 0.4729
#> X2~~X2 1.0000 0.0000 20000 1.0000 1.0000
#> X2~~X3 0.3743 0.0615 20000 0.2481 0.4909
#> X3~~X3 1.0000 0.0000 20000 1.0000 1.0000
#> rsq1 0.0572 0.0322 20000 0.0114 0.1339
#> rsqbar1 0.0619 0.0320 20000 0.0163 0.1382
#> rsq2 0.2572 0.0530 20000 0.1649 0.3709
#> rsqbar2 0.2534 0.0532 20000 0.1607 0.3677