Benchmark: Comparing the Monte Carlo Method with Nonparametric Bootstrapping
Ivan Jacob Agaloos Pesigan
2025-10-19
Source:vignettes/benchmark-complete.Rmd
benchmark-complete.RmdWe compare the Monte Carlo (MC) method with nonparametric bootstrapping (NB) using the simple mediation model with complete data. One advantage of MC over NB is speed. This is because the model is only fitted once in MC whereas it is fitted many times in NB.
Model Specification
The indirect effect is defined by the product of the slopes of paths
X to M labeled as a and
M to Y labeled as b. In this
example, we are interested in the confidence intervals of
indirect defined as the product of a and
b using the := operator in the
lavaan model syntax.
model <- "
Y ~ cp * X + b * M
M ~ a * X
X ~~ X
indirect := a * b
direct := cp
total := cp + (a * b)
"Model Fitting
We can now fit the model using the sem() function from
lavaan.
fit <- sem(data = df, model = model)Monte Carlo Confidence Intervals
The fit lavaan object can then be passed to
the MC() function from semmcci to generate
Monte Carlo confidence intervals.
MC(fit, R = 100L, alpha = 0.05)
#> Monte Carlo Confidence Intervals
#> est se R 2.5% 97.5%
#> cp 0.2333 0.0296 100 0.1806 0.2903
#> b 0.5082 0.0279 100 0.4555 0.5527
#> a 0.4820 0.0280 100 0.4220 0.5301
#> X~~X 1.0590 0.0426 100 0.9751 1.1296
#> Y~~Y 0.5462 0.0231 100 0.5064 0.5959
#> M~~M 0.7527 0.0337 100 0.7024 0.8208
#> indirect 0.2449 0.0179 100 0.2058 0.2738
#> direct 0.2333 0.0296 100 0.1806 0.2903
#> total 0.4782 0.0295 100 0.4162 0.5283Nonparametric Bootstrap Confidence Intervals
Nonparametric bootstrap confidence intervals can be generated in
lavaan using the following.
parameterEstimates(
sem(
data = df,
model = model,
se = "bootstrap",
bootstrap = 100L
)
)
#> lhs op rhs label est se z pvalue ci.lower ci.upper
#> 1 Y ~ X cp 0.233 0.025 9.395 0 0.183 0.278
#> 2 Y ~ M b 0.508 0.028 18.057 0 0.454 0.568
#> 3 M ~ X a 0.482 0.026 18.550 0 0.433 0.535
#> 4 X ~~ X 1.059 0.046 23.224 0 0.969 1.161
#> 5 Y ~~ Y 0.546 0.023 23.640 0 0.508 0.593
#> 6 M ~~ M 0.753 0.033 23.131 0 0.692 0.814
#> 7 indirect := a*b indirect 0.245 0.020 12.443 0 0.209 0.289
#> 8 direct := cp direct 0.233 0.025 9.395 0 0.183 0.278
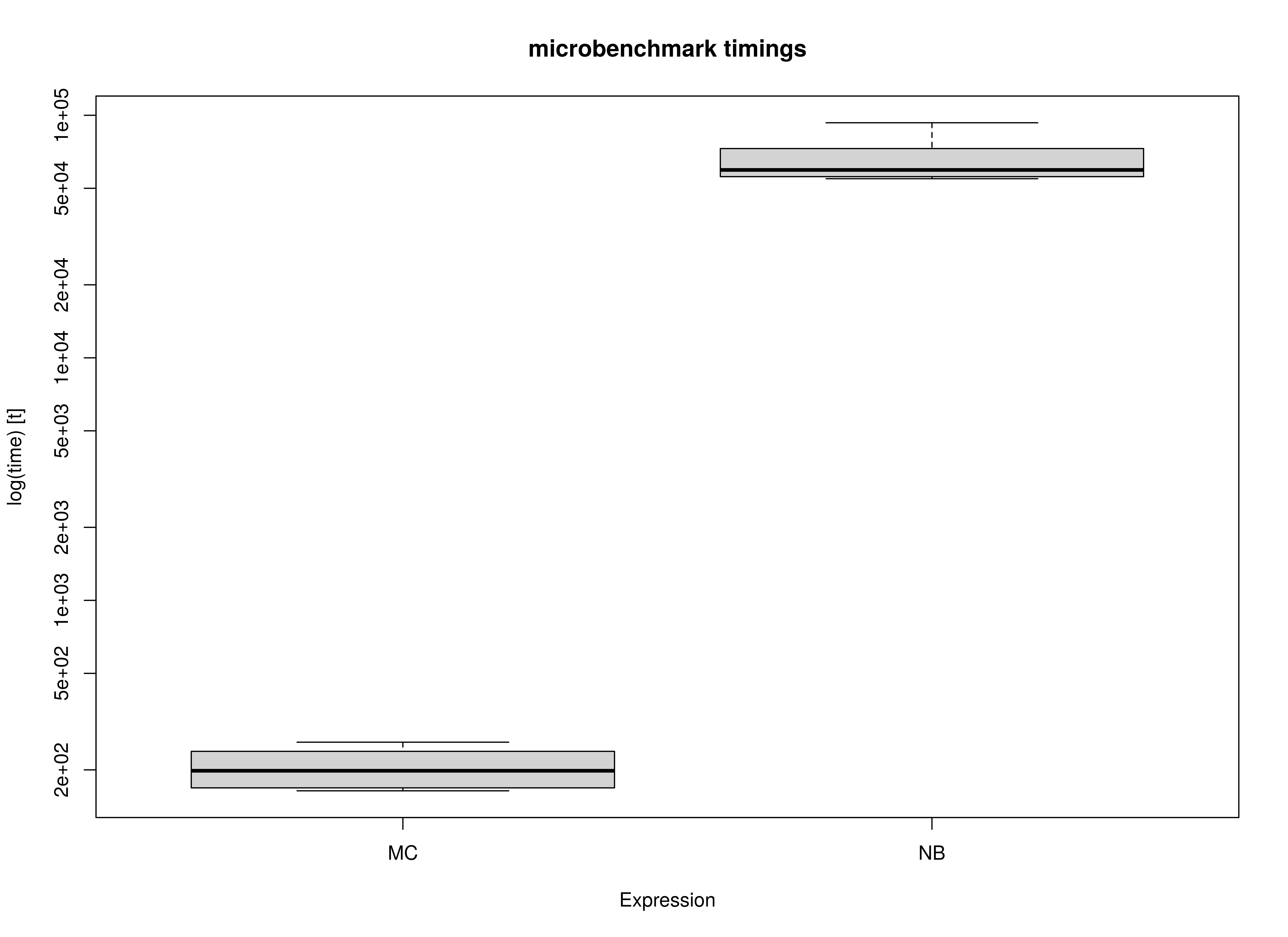
#> 9 total := cp+(a*b) total 0.478 0.027 17.966 0 0.418 0.518Benchmark
Arguments
| Variables | Values | Notes |
|---|---|---|
| R | 1000 | Number of Monte Carlo replications. |
| B | 1000 | Number of bootstrap samples. |
benchmark_complete_01 <- microbenchmark(
MC = {
fit <- sem(
data = df,
model = model
)
MC(
fit,
R = R,
decomposition = "chol",
pd = FALSE
)
},
NB = sem(
data = df,
model = model,
se = "bootstrap",
bootstrap = B
),
times = 10
)Summary of Benchmark Results
summary(benchmark_complete_01, unit = "ms")
#> expr min lq mean median uq max
#> 1 MC 71.92246 72.26498 75.40017 73.03176 74.11578 97.32907
#> 2 NB 13134.40864 13199.61712 13220.57029 13210.41936 13228.86796 13318.20412
#> neval
#> 1 10
#> 2 10Summary of Benchmark Results Relative to the Faster Method
summary(benchmark_complete_01, unit = "relative")
#> expr min lq mean median uq max neval
#> 1 MC 1.000 1.0000 1.0000 1.0000 1.0000 1.0000 10
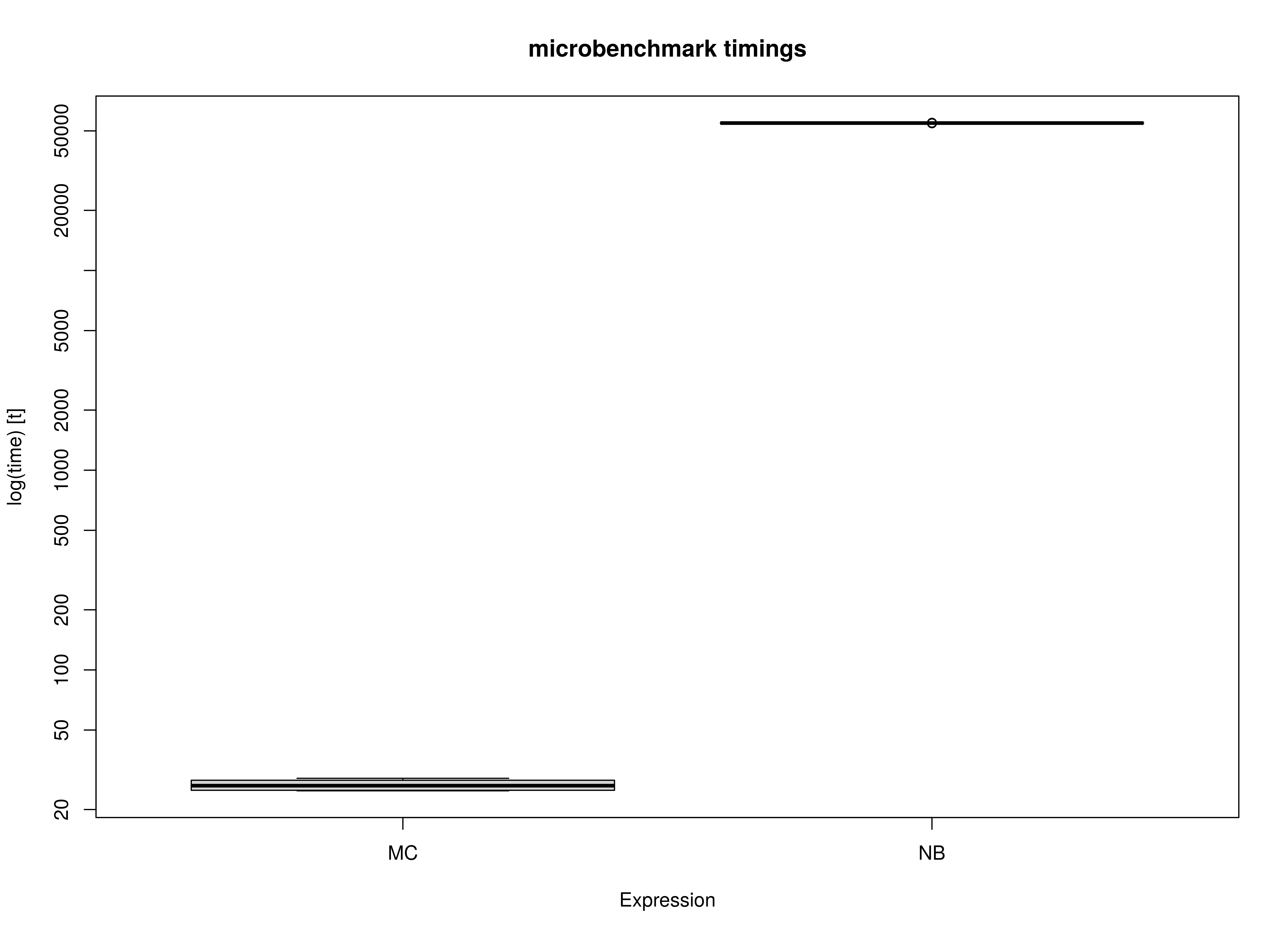
#> 2 NB 182.619 182.6558 175.3387 180.8859 178.4892 136.8369 10Benchmark - Monte Carlo Method with Precalculated Estimates
fit <- sem(
data = df,
model = model
)
benchmark_complete_02 <- microbenchmark(
MC = MC(
fit,
R = R,
decomposition = "chol",
pd = FALSE
),
NB = sem(
data = df,
model = model,
se = "bootstrap",
bootstrap = B
),
times = 10
)Summary of Benchmark Results
summary(benchmark_complete_02, unit = "ms")
#> expr min lq mean median uq max
#> 1 MC 21.42988 21.53643 22.90902 21.91124 25.2187 26.11223
#> 2 NB 13142.92047 13147.53284 13162.94078 13153.17865 13171.4634 13204.09618
#> neval
#> 1 10
#> 2 10Summary of Benchmark Results Relative to the Faster Method
summary(benchmark_complete_02, unit = "relative")
#> expr min lq mean median uq max neval
#> 1 MC 1.000 1.0000 1.0000 1.0000 1.0000 1.0000 10
#> 2 NB 613.299 610.4787 574.5745 600.2937 522.2896 505.6671 10