Fit the Discrete-Time Vector Autoregressive Model By ID (Stable Reciprocal Regulation)
Ivan Jacob Agaloos Pesigan
2026-01-08
Source:vignettes/stable-reciprocal-regulation-1000.Rmd
stable-reciprocal-regulation-1000.RmdDynamics Description
The Stable Reciprocal Regulation process represents a bivariate dynamic system in which two latent psychological constructs—such as positive and negative affect—mutually influence each other over time. Each construct shows moderate self-regulation (autoregressive effects) and mild opposing cross-effects, reflecting an equilibrium-seeking mechanism characteristic of emotional balance.
Individuals vary in their self-regulatory tendencies and in the strength of these antagonistic couplings. At the population level, the transition matrix indicates that increases in one construct are followed by slight decreases in the other, producing a stable, damped oscillatory pattern around individual equilibrium points. The process noise covariance allows for small correlated disturbances, while measurement errors are assumed to be minimal and symmetric across indicators.
This dynamic pattern captures a psychologically plausible process of reciprocal inhibition—where short-term fluctuations in one system component (e.g., positive affect) are naturally counteracted by adjustments in its counterpart (e.g., negative affect), leading to emotional homeostasis over time.
Model
The measurement model is given by where , , and are random variables and , , and are model parameters. represents a vector of observed random variables, a vector of latent random variables, and a vector of random measurement errors, at time and individual . denotes a vector of person-specific intercepts while denotes a matrix of factor loadings, and the covariance matrix of that is invariant across individuals. In this model, is an identity matrix and is a symmetric matrix.
The dynamic structure is given by where , , and are random variables, and , and are model parameters. Here, is a vector of latent variables at time and individual , represents a vector of latent variables at time and individual , and represents a vector of dynamic noise at time and individual . is a matrix of autoregression and cross regression coefficients for individual , and the covariance matrix of that is invariant across all individuals. In this model, is a symmetric matrix.
Data Generation
Notation
Let be the number of time points and be the number of individuals. We simulate a total of time points per individual, discarding the first as burn-in. The analysis uses the final measurement occasions.
Let the measurement model intercept vector be normally distributed with the following means
and covariance matrix
Let the factor loadings matrix be given by
Let the measurement error covariance matrix be given by
Let the initial condition be given by
and are functions of and .
Let the transition matrix be normally distributed with the following means
and covariance matrix
Let the intercept vector be fixed to a zero vector.
The SimNuN and SimBetaN functions from the
simStateSpace package generate random intercept vectors and
transition matrices from the multivariate normal distribution. Note that
the SimBetaN function generates transition matrices that
are weakly stationary with an option to set lower and upper bounds.
Let the dynamic process noise be given by
R Function Arguments
n
#> [1] 1000
time
#> [1] 11000
burnin
#> [1] 10000
# first mu0 in the list of length n
mu0[[1]]
#> [1] 0 0
# first sigma0 in the list of length n
sigma0[[1]]
#> [,1] [,2]
#> [1,] 0.3230830 -0.1220195
#> [2,] -0.1220195 0.5527409
# first sigma0_l in the list of length n
sigma0_l[[1]] # sigma0_l <- t(chol(sigma0))
#> [,1] [,2]
#> [1,] 0.5684039 0.0000000
#> [2,] -0.2146704 0.7117988
alpha
#> [[1]]
#> [1] 0 0
# first beta in the list of length n
beta[[1]]
#> [,1] [,2]
#> [1,] 0.6248343 0.02174497
#> [2,] -0.1054576 0.79427204
# first psi in the list of length n
psi[[1]]
#> [,1] [,2]
#> [1,] 0.20 -0.05
#> [2,] -0.05 0.18
psi_l[[1]] # psi_l <- t(chol(psi))
#> [,1] [,2]
#> [1,] 0.4472136 0.0000000
#> [2,] -0.1118034 0.4092676
# first nu in the list of length n
nu[[1]]
#> [1] 0.5230419 -0.1853803
lambda
#> [[1]]
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
# first theta in the list of length n
theta[[1]]
#> [,1] [,2]
#> [1,] 0.5 0.0
#> [2,] 0.0 0.5
theta_l[[1]] # theta_l <- t(chol(theta))
#> [,1] [,2]
#> [1,] 0.7071068 0.0000000
#> [2,] 0.0000000 0.7071068Visualizing the Dynamics Without Process Noise and Measurement Error (n = 5 with Different Initial Condition)
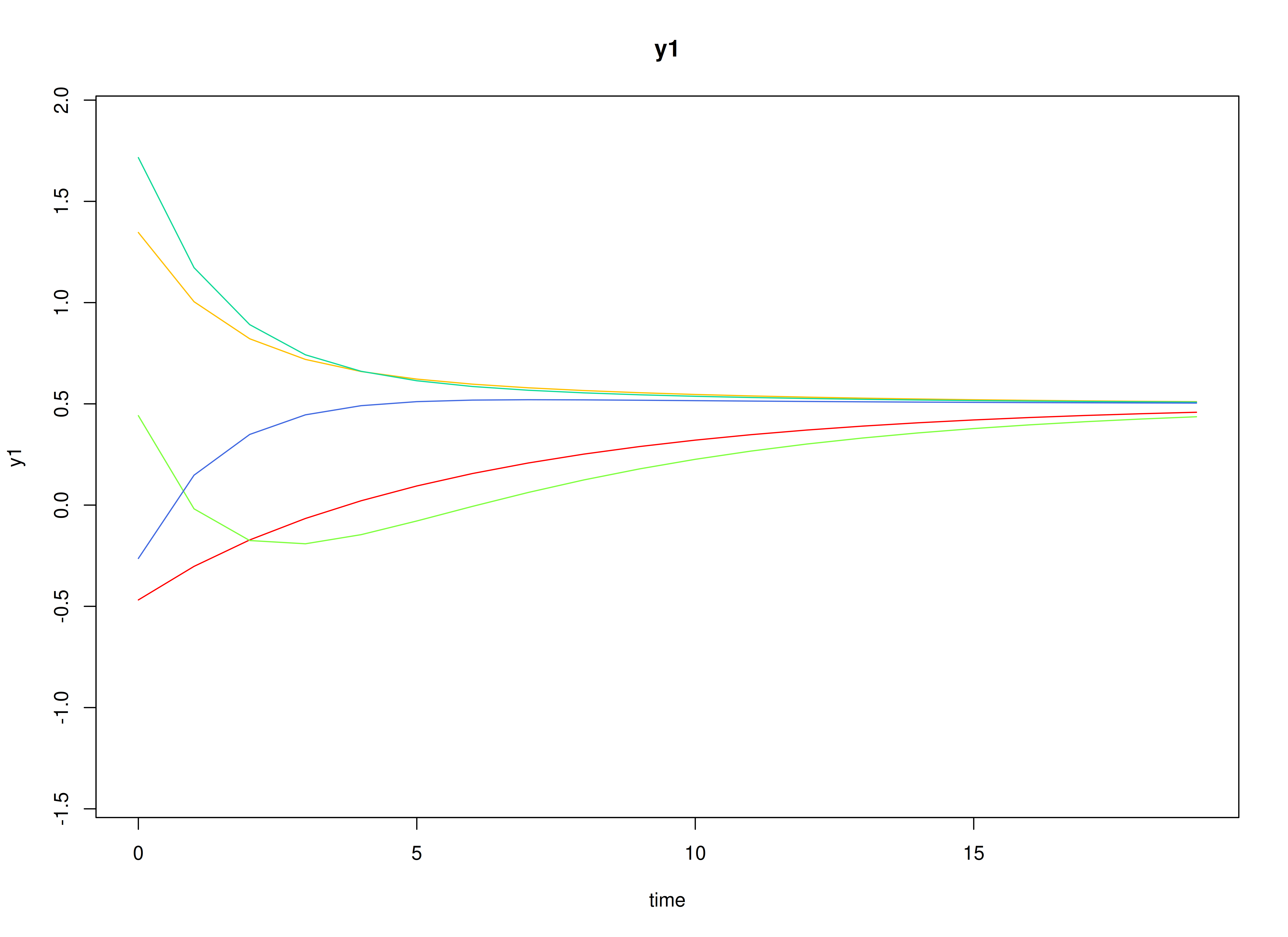
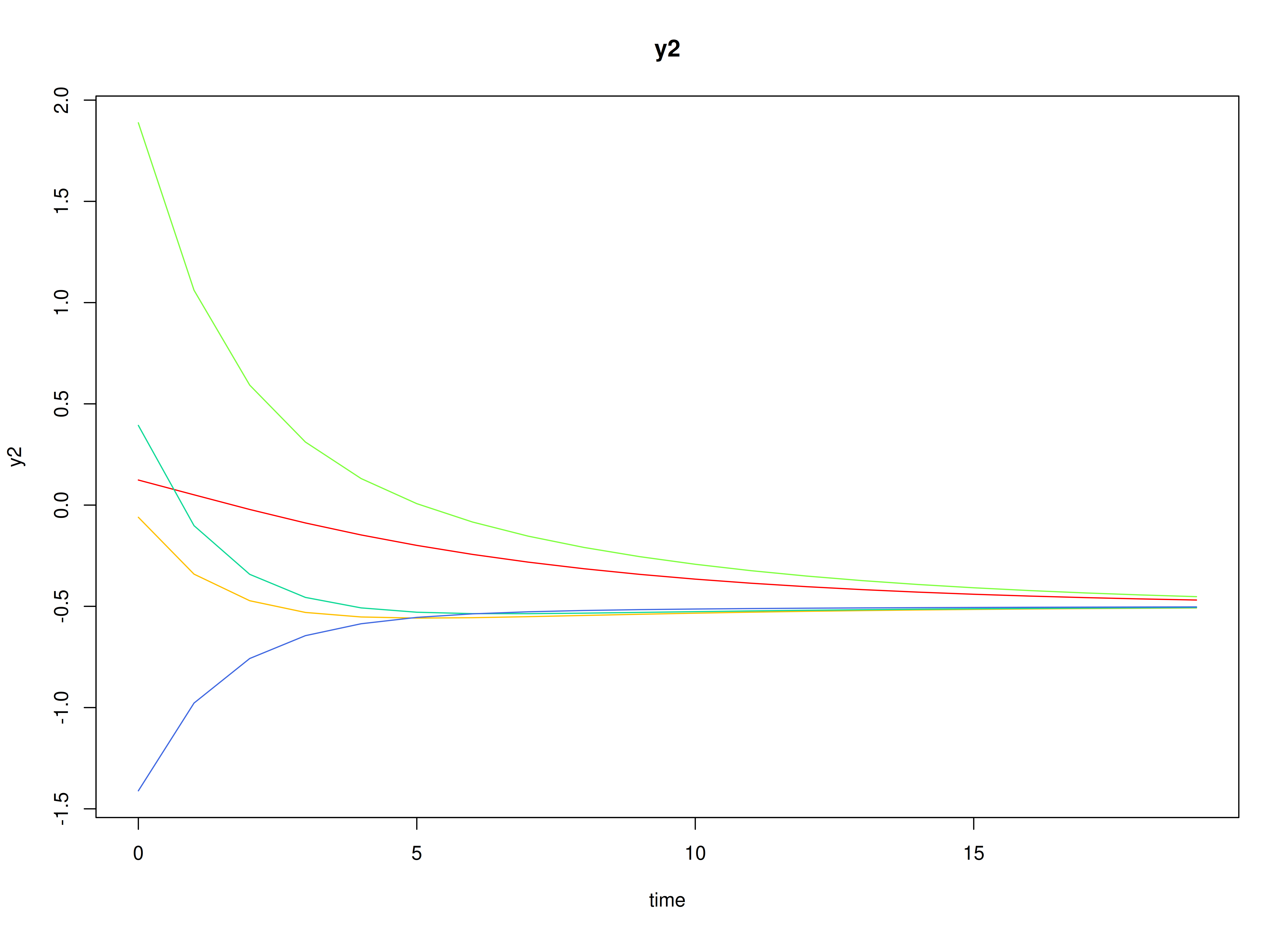
Using the SimSSMIVary Function from the
simStateSpace Package to Simulate Data
library(simStateSpace)
sim <- SimSSMIVary(
n = n,
time = time,
mu0 = mu0,
sigma0_l = sigma0_l,
alpha = alpha,
beta = beta,
psi_l = psi_l,
nu = nu,
lambda = lambda,
theta_l = theta_l
)
data <- as.data.frame(sim, burnin = burnin)
head(data)
#> id time y1 y2
#> 1 1 0 -0.39108331 -0.3009975
#> 2 1 1 0.03198217 -0.9235198
#> 3 1 2 0.90362186 0.2655117
#> 4 1 3 -0.32032150 0.0151437
#> 5 1 4 -0.26009422 -0.4996087
#> 6 1 5 1.12660279 -0.7440601
plot(sim, burnin = burnin)
Model Fitting
The FitDTVARMxID function fits a DT-VAR model on each
individual
.
To set up the estimation, we first provide starting
values for each parameter matrix.
Autoregressive Parameters (beta)
We initialize the autoregressive coefficient matrix with the true values used in simulation.
beta_values <- betaLDL’-parameterized covariance matrices
Covariances such as psi and theta are
estimated using the LDL’ decomposition of a positive definite covariance
matrix. The decomposition expresses a covariance matrix
as
where:
-
is a strictly lower-triangular matrix of free parameters
(
l_mat_strict),
-
is the identity matrix,
-
is an unconstrained vector,
- ensures strictly positive diagonal entries.
The LDL() function extracts this decomposition from a
positive definite covariance matrix. It returns:
-
d_uc: unconstrained diagonal parameters, equal toInvSoftplus(d_vec),
-
d_vec: diagonal entries, equal toSoftplus(d_uc),
-
l_mat_strict: the strictly lower-triangular factor.
sigma <- matrix(
data = c(1.0, 0.5, 0.5, 1.0),
nrow = 2,
ncol = 2
)
ldl_sigma <- LDL(sigma)
d_uc <- ldl_sigma$d_uc
l_mat_strict <- ldl_sigma$l_mat_strict
I <- diag(2)
sigma_reconstructed <- (l_mat_strict + I) %*% diag(log1p(exp(d_uc)), 2) %*% t(l_mat_strict + I)
sigma_reconstructed
#> [,1] [,2]
#> [1,] 1.0 0.5
#> [2,] 0.5 1.0Process Noise Covariance Matrix (psi)
Starting values for the process noise covariance matrix are given below, with corresponding LDL’ parameters.
psi_values <- psi[[1]]
ldl_psi_values <- LDL(psi_values)
psi_d_values <- ldl_psi_values$d_uc
psi_l_values <- ldl_psi_values$l_mat_strict
psi_d_values
#> [1] -1.507772 -1.701853
psi_l_values
#> [,1] [,2]
#> [1,] 0.00 0
#> [2,] -0.25 0Measurement Error Covariance Matrix (theta)
Starting values for the measurement error covariance matrix are given below, with corresponding LDL’ parameters.
theta_values <- theta[[1]]
ldl_theta_values <- LDL(theta_values)
theta_d_values <- ldl_theta_values$d_uc
theta_l_values <- ldl_theta_values$l_mat_strict
theta_d_values
#> [1] -0.4327521 -0.4327521
theta_l_values
#> [,1] [,2]
#> [1,] 0 0
#> [2,] 0 0Initial mean vector (mu_0) and covariance matrix
(sigma_0)
The initial mean vector
and covariance matrix
are fixed using mu0 and sigma0.
mu0_values <- mu0
FitDTVARMxID
fit <- FitDTVARMxID(
data = data,
observed = c("y1", "y2"),
id = "id",
beta_values = beta_values,
psi_d_values = psi_d_values,
psi_l_values = psi_l_values,
nu_values = nu_values,
theta_d_values = theta_d_values,
mu0_values = mu0_values,
sigma0_d_values = sigma0_d_values,
sigma0_l_values = sigma0_l_values,
ncores = parallel::detectCores()
)Parameter estimates
head(summary(fit))
#> beta_1_1 beta_2_1 beta_1_2 beta_2_2
#> FitDTVARMxID_DTVAR_ID1.Rds 0.6374979 -0.17348371 0.02575771 0.7937547
#> FitDTVARMxID_DTVAR_ID2.Rds 0.7542477 -0.16899466 -0.10221295 0.8516929
#> FitDTVARMxID_DTVAR_ID3.Rds 0.6172794 -0.05090336 -0.13723502 0.6130297
#> FitDTVARMxID_DTVAR_ID4.Rds 0.6753522 -0.09814573 -0.15948403 0.7858947
#> FitDTVARMxID_DTVAR_ID5.Rds 0.7020813 -0.12182501 -0.11240540 0.7177757
#> FitDTVARMxID_DTVAR_ID6.Rds 0.3761940 -0.11338911 -0.10006777 0.4990226
#> nu_1_1 nu_2_1 psi_l_2_1 psi_d_1_1
#> FitDTVARMxID_DTVAR_ID1.Rds 0.5550406 -0.12508171 -0.1439181 -1.719023
#> FitDTVARMxID_DTVAR_ID2.Rds 0.4302223 -0.36358145 -0.2959200 -1.554567
#> FitDTVARMxID_DTVAR_ID3.Rds 0.2258173 0.09403236 -0.2617573 -1.146391
#> FitDTVARMxID_DTVAR_ID4.Rds 0.1153293 -0.01231006 -0.4056990 -1.776335
#> FitDTVARMxID_DTVAR_ID5.Rds 0.7902069 -0.74676206 -0.2975987 -1.410096
#> FitDTVARMxID_DTVAR_ID6.Rds 0.9108391 -0.73651068 -0.3186177 -1.061309
#> psi_d_2_1 theta_d_1_1 theta_d_2_1
#> FitDTVARMxID_DTVAR_ID1.Rds -2.0863281 -0.5006095 -0.3650390
#> FitDTVARMxID_DTVAR_ID2.Rds -1.4709971 -0.3724268 -0.4119888
#> FitDTVARMxID_DTVAR_ID3.Rds -1.9314857 -0.4768679 -0.1812760
#> FitDTVARMxID_DTVAR_ID4.Rds -1.7841345 -0.4259500 -0.3055352
#> FitDTVARMxID_DTVAR_ID5.Rds -1.3809979 -0.5156890 -0.6252657
#> FitDTVARMxID_DTVAR_ID6.Rds -0.9290789 -0.5957023 -0.8837091Proportion of converged cases
converged(
fit,
theta_tol = 0.01,
prop = TRUE
)
#> [1] 0.937Fixed-Effect Meta-Analysis of Measurement Error
When fitting DT-VAR models per person, separating process noise () from measurement error () can be unstable for some individuals. To stabilize inference, we first pool the person-level estimates from only the converged fits using a fixed-effect meta-analysis. This yields a high-precision estimate of the common measurement-error covariance that we will then hold fixed in a second pass of model fitting.
What the code does: - Selects individuals that converged and whose
diagonals exceed a small threshold (theta_tol), filtering
out near-zero or ill-conditioned solutions. - Extracts each person’s
LDL’ diagonal parameters for
and their sampling covariance matrices. - Computes the
inverse-variance-weighted pooled estimate (fixed effect), returning it
on the same LDL’ parameterization used by
FitDTVARMxID().
library(metaVAR)
fixed_theta <- MetaVARMx(
fit,
random = FALSE, # TRUE by default
effects = FALSE, # TRUE by default
cov_meas = TRUE, # FALSE by default
theta_tol = 0.01,
ncores = parallel::detectCores()
)You can read summary(fixed_theta) as providing the
pooled (fixed) measurement-error scale that is common across persons. If
individual instruments truly share the same reliability structure,
fixing
to this pooled value improves stability and often reduces bias in the
dynamic parameters.
Note: Fixed-effect pooling assumes a common across individuals.
coef(fixed_theta)
#> alpha_1_1 alpha_2_1
#> -0.3964559 -0.3971429
summary(fixed_theta)
#> [1] 0
#> Call:
#> MetaVARMx(object = fit, random = FALSE, effects = FALSE, cov_meas = TRUE,
#> theta_tol = 0.01, ncores = parallel::detectCores())
#>
#> CI type = "normal"
#> est se z p 2.5% 97.5%
#> alpha[1,1] -0.3965 0.0041 -95.6497 0 -0.4046 -0.3883
#> alpha[2,1] -0.3971 0.0044 -90.3312 0 -0.4058 -0.3885
theta_d_values <- coef(fixed_theta)Refit the model with fixed measurement error covariance matrix
We refit the individual models using the pooled as a fixed measurement-error covariance matrix.
fit <- FitDTVARMxID(
data = data,
observed = c("y1", "y2"),
id = "id",
beta_values = beta_values,
psi_d_values = psi_d_values,
psi_l_values = psi_l_values,
nu_values = nu_values,
theta_fixed = TRUE,
theta_d_values = theta_d_values,
mu0_values = mu0_values,
sigma0_d_values = sigma0_d_values,
sigma0_l_values = sigma0_l_values,
ncores = parallel::detectCores()
)With fixed, the re-estimation focuses on the dynamic structure (, ) and intercepts (). In practice, this often increases the proportion of converged fits and yields more stable cross-lag estimates.
Proportion of converged cases
converged(
fit,
prop = TRUE
)
#> [1] 1Random-Effects Meta-Analysis of Person-Specific Dynamics and Means
Having stabilized , we synthesize the person-specific estimates to recover population-level effects and their between-person variability. We use a random-effects model so the pooled mean reflects both within-person estimation uncertainty and between-person heterogeneity.
random <- MetaVARMx(
fit,
effects = TRUE,
int_meas = TRUE,
robust_v = FALSE,
robust = TRUE,
ncores = parallel::detectCores()
)
summary(random)
#> [1] 0
#> Call:
#> MetaVARMx(object = fit, effects = TRUE, int_meas = TRUE, robust_v = FALSE,
#> robust = TRUE, ncores = parallel::detectCores())
#>
#> CI type = "normal"
#> est se z p 2.5% 97.5%
#> alpha[1,1] 0.7075 0.0041 172.9477 0.0000 0.6995 0.7155
#> alpha[2,1] -0.1250 0.0041 -30.5559 0.0000 -0.1330 -0.1170
#> alpha[3,1] -0.1671 0.0038 -43.4118 0.0000 -0.1746 -0.1596
#> alpha[4,1] 0.6760 0.0039 175.2079 0.0000 0.6684 0.6836
#> alpha[5,1] 0.5365 0.0109 49.3161 0.0000 0.5152 0.5578
#> alpha[6,1] -0.5231 0.0102 -51.1214 0.0000 -0.5432 -0.5030
#> tau_sqr[1,1] 0.0118 0.0007 16.7242 0.0000 0.0104 0.0132
#> tau_sqr[2,1] 0.0074 0.0006 13.2017 0.0000 0.0063 0.0085
#> tau_sqr[3,1] -0.0002 0.0005 -0.5390 0.5899 -0.0012 0.0007
#> tau_sqr[4,1] 0.0003 0.0005 0.5992 0.5491 -0.0006 0.0012
#> tau_sqr[5,1] -0.0030 0.0014 -2.1390 0.0324 -0.0057 -0.0002
#> tau_sqr[6,1] 0.0006 0.0013 0.4726 0.6365 -0.0019 0.0031
#> tau_sqr[2,2] 0.0122 0.0007 16.7491 0.0000 0.0108 0.0136
#> tau_sqr[3,2] -0.0019 0.0004 -4.2850 0.0000 -0.0027 -0.0010
#> tau_sqr[4,2] 0.0002 0.0005 0.3434 0.7313 -0.0007 0.0011
#> tau_sqr[5,2] -0.0015 0.0014 -1.0922 0.2747 -0.0041 0.0012
#> tau_sqr[6,2] -0.0001 0.0013 -0.0495 0.9605 -0.0026 0.0024
#> tau_sqr[3,3] 0.0087 0.0006 14.3251 0.0000 0.0075 0.0098
#> tau_sqr[4,3] 0.0038 0.0004 8.7142 0.0000 0.0030 0.0047
#> tau_sqr[5,3] -0.0004 0.0013 -0.2794 0.7800 -0.0028 0.0021
#> tau_sqr[6,3] 0.0006 0.0012 0.5479 0.5838 -0.0017 0.0030
#> tau_sqr[4,4] 0.0089 0.0006 14.9336 0.0000 0.0078 0.0101
#> tau_sqr[5,4] 0.0019 0.0013 1.4952 0.1349 -0.0006 0.0044
#> tau_sqr[6,4] -0.0009 0.0012 -0.7131 0.4758 -0.0032 0.0015
#> tau_sqr[5,5] 0.1085 0.0053 20.5695 0.0000 0.0981 0.1188
#> tau_sqr[6,5] -0.0527 0.0039 -13.3413 0.0000 -0.0604 -0.0449
#> tau_sqr[6,6] 0.0976 0.0046 20.9925 0.0000 0.0885 0.1067
#> i_sqr[1,1] 0.9196 0.0044 208.0888 0.0000 0.9110 0.9283
#> i_sqr[2,1] 0.9305 0.0038 242.9527 0.0000 0.9230 0.9380
#> i_sqr[3,1] 0.9489 0.0026 370.6043 0.0000 0.9439 0.9539
#> i_sqr[4,1] 0.9529 0.0023 409.6193 0.0000 0.9483 0.9575
#> i_sqr[5,1] 0.9698 0.0014 680.2652 0.0000 0.9670 0.9726
#> i_sqr[6,1] 0.9790 0.0010 1013.1868 0.0000 0.9771 0.9809Normal Theory Confidence Intervals
confint(random, level = 0.95, lb = FALSE)
#> 2.5 % 97.5 %
#> alpha[1,1] 0.6994614092 0.7154967018
#> alpha[2,1] -0.1330007621 -0.1169669424
#> alpha[3,1] -0.1746422004 -0.1595538581
#> alpha[4,1] 0.6684393011 0.6835634858
#> alpha[5,1] 0.5151558123 0.5577980855
#> alpha[6,1] -0.5431570185 -0.5030462320
#> tau_sqr[1,1] 0.0104438463 0.0132167001
#> tau_sqr[2,1] 0.0062783696 0.0084676030
#> tau_sqr[3,1] -0.0011552849 0.0006569005
#> tau_sqr[4,1] -0.0006136405 0.0011540327
#> tau_sqr[5,1] -0.0056831546 -0.0002482561
#> tau_sqr[6,1] -0.0018774036 0.0030705119
#> tau_sqr[2,2] 0.0107645166 0.0136177077
#> tau_sqr[3,2] -0.0027460049 -0.0010223533
#> tau_sqr[4,2] -0.0007418905 0.0010571365
#> tau_sqr[5,2] -0.0041289345 0.0011738603
#> tau_sqr[6,2] -0.0025724749 0.0024456554
#> tau_sqr[3,3] 0.0074678486 0.0098352700
#> tau_sqr[4,3] 0.0029800188 0.0047095283
#> tau_sqr[5,3] -0.0028236486 0.0021191291
#> tau_sqr[6,3] -0.0016635329 0.0029543666
#> tau_sqr[4,4] 0.0077518786 0.0100940669
#> tau_sqr[5,4] -0.0005863408 0.0043588855
#> tau_sqr[6,4] -0.0032265027 0.0015050996
#> tau_sqr[5,5] 0.0981185096 0.1187862527
#> tau_sqr[6,5] -0.0603980479 -0.0449250395
#> tau_sqr[6,6] 0.0884843535 0.1067084946
#> i_sqr[1,1] 0.9109567775 0.9282803347
#> i_sqr[2,1] 0.9229764371 0.9379893399
#> i_sqr[3,1] 0.9439014198 0.9539382644
#> i_sqr[4,1] 0.9483426818 0.9574616569
#> i_sqr[5,1] 0.9669664359 0.9725545377
#> i_sqr[6,1] 0.9770800388 0.9808675997
confint(random, level = 0.99, lb = FALSE)
#> 0.5 % 99.5 %
#> alpha[1,1] 0.6969420822 0.7180160288
#> alpha[2,1] -0.1355198577 -0.1144478468
#> alpha[3,1] -0.1770127508 -0.1571833077
#> alpha[4,1] 0.6660631194 0.6859396674
#> alpha[5,1] 0.5084562256 0.5644976722
#> alpha[6,1] -0.5494588798 -0.4967443707
#> tau_sqr[1,1] 0.0100081995 0.0136523470
#> tau_sqr[2,1] 0.0059344161 0.0088115565
#> tau_sqr[3,1] -0.0014399999 0.0009416155
#> tau_sqr[4,1] -0.0008913621 0.0014317543
#> tau_sqr[5,1] -0.0065370391 0.0006056283
#> tau_sqr[6,1] -0.0026547775 0.0038478857
#> tau_sqr[2,2] 0.0103162478 0.0140659765
#> tau_sqr[3,2] -0.0030168102 -0.0007515480
#> tau_sqr[4,2] -0.0010245382 0.0013397842
#> tau_sqr[5,2] -0.0049620640 0.0020069898
#> tau_sqr[6,2] -0.0033608803 0.0032340609
#> tau_sqr[3,3] 0.0070958997 0.0102072189
#> tau_sqr[4,3] 0.0027082931 0.0049812540
#> tau_sqr[5,3] -0.0036002153 0.0028956958
#> tau_sqr[6,3] -0.0023890575 0.0036798912
#> tau_sqr[4,4] 0.0073838941 0.0104620514
#> tau_sqr[5,4] -0.0013632922 0.0051358368
#> tau_sqr[6,4] -0.0039698914 0.0022484882
#> tau_sqr[5,5] 0.0948713718 0.1220333904
#> tau_sqr[6,5] -0.0628290337 -0.0424940537
#> tau_sqr[6,6] 0.0856211334 0.1095717147
#> i_sqr[1,1] 0.9082350494 0.9310020628
#> i_sqr[2,1] 0.9206177391 0.9403480378
#> i_sqr[3,1] 0.9423245172 0.9555151670
#> i_sqr[4,1] 0.9469099870 0.9588943517
#> i_sqr[5,1] 0.9660884815 0.9734324922
#> i_sqr[6,1] 0.9764849698 0.9814626687Robust Confidence Intervals
confint(random, level = 0.95, lb = FALSE, robust = TRUE)
#> 2.5 % 97.5 %
#> alpha[1,1] 0.6992133279 0.7157447831
#> alpha[2,1] -0.1333447988 -0.1166229057
#> alpha[3,1] -0.1749502077 -0.1592458509
#> alpha[4,1] 0.6682576101 0.6837451767
#> alpha[5,1] 0.5150965585 0.5578573393
#> alpha[6,1] -0.5431395308 -0.5030637197
#> tau_sqr[1,1] 0.0104375090 0.0132230374
#> tau_sqr[2,1] 0.0061688059 0.0085771666
#> tau_sqr[3,1] -0.0011611014 0.0006627169
#> tau_sqr[4,1] -0.0006469710 0.0011873631
#> tau_sqr[5,1] -0.0057418008 -0.0001896100
#> tau_sqr[6,1] -0.0018486313 0.0030417396
#> tau_sqr[2,2] 0.0107500822 0.0136321420
#> tau_sqr[3,2] -0.0027171663 -0.0010511919
#> tau_sqr[4,2] -0.0007641090 0.0010793550
#> tau_sqr[5,2] -0.0040036368 0.0010485626
#> tau_sqr[6,2] -0.0025869404 0.0024601210
#> tau_sqr[3,3] 0.0075585681 0.0097445506
#> tau_sqr[4,3] 0.0029447682 0.0047447789
#> tau_sqr[5,3] -0.0027769018 0.0020723822
#> tau_sqr[6,3] -0.0015945791 0.0028854128
#> tau_sqr[4,4] 0.0077672041 0.0100787414
#> tau_sqr[5,4] -0.0006376927 0.0044102374
#> tau_sqr[6,4] -0.0032960568 0.0015746537
#> tau_sqr[5,5] 0.0987007591 0.1182040031
#> tau_sqr[6,5] -0.0603698940 -0.0449531934
#> tau_sqr[6,6] 0.0889572795 0.1062355685
#> i_sqr[1,1] 0.9109171845 0.9283199276
#> i_sqr[2,1] 0.9228914259 0.9380743511
#> i_sqr[3,1] 0.9437745418 0.9540651424
#> i_sqr[4,1] 0.9484093321 0.9573950067
#> i_sqr[5,1] 0.9671239674 0.9723970063
#> i_sqr[6,1] 0.9771594831 0.9807881554
confint(random, level = 0.99, lb = FALSE, robust = TRUE)
#> 0.5 % 99.5 %
#> alpha[1,1] 0.6966160480 0.7183420629
#> alpha[2,1] -0.1359719986 -0.1139957058
#> alpha[3,1] -0.1774175409 -0.1567785176
#> alpha[4,1] 0.6658243371 0.6861784497
#> alpha[5,1] 0.5083783529 0.5645755449
#> alpha[6,1] -0.5494358970 -0.4967673535
#> tau_sqr[1,1] 0.0099998708 0.0136606757
#> tau_sqr[2,1] 0.0057904251 0.0089555475
#> tau_sqr[3,1] -0.0014476440 0.0009492595
#> tau_sqr[4,1] -0.0009351657 0.0014755579
#> tau_sqr[5,1] -0.0066141132 0.0006827025
#> tau_sqr[6,1] -0.0026169643 0.0038100726
#> tau_sqr[2,2] 0.0102972778 0.0140849464
#> tau_sqr[3,2] -0.0029789098 -0.0007894484
#> tau_sqr[4,2] -0.0010537382 0.0013689842
#> tau_sqr[5,2] -0.0047973949 0.0018423207
#> tau_sqr[6,2] -0.0033798912 0.0032530718
#> tau_sqr[3,3] 0.0072151253 0.0100879933
#> tau_sqr[4,3] 0.0026619660 0.0050275811
#> tau_sqr[5,3] -0.0035387795 0.0028342600
#> tau_sqr[6,3] -0.0022984368 0.0035892705
#> tau_sqr[4,4] 0.0074040352 0.0104419103
#> tau_sqr[5,4] -0.0014307800 0.0052033247
#> tau_sqr[6,4] -0.0040613008 0.0023398977
#> tau_sqr[5,5] 0.0956365774 0.1212681849
#> tau_sqr[6,5] -0.0627920332 -0.0425310542
#> tau_sqr[6,6] 0.0862426636 0.1089501844
#> i_sqr[1,1] 0.9081830154 0.9310540967
#> i_sqr[2,1] 0.9205060155 0.9404597615
#> i_sqr[3,1] 0.9421577712 0.9556819130
#> i_sqr[4,1] 0.9469975803 0.9588067585
#> i_sqr[5,1] 0.9662955130 0.9732254607
#> i_sqr[6,1] 0.9765893774 0.9813582611Profile-Likelihood Confidence Intervals
confint(random, level = 0.95, lb = TRUE)
#> 2.5 % 97.5 %
#> alpha[1,1] 0.6993725548 0.7154363329
#> alpha[2,1] -0.1330920923 -0.1169835645
#> alpha[3,1] -0.1747435699 -0.1596002873
#> alpha[4,1] 0.6683361945 0.6835338180
#> alpha[5,1] 0.5150555461 0.5578187359
#> alpha[6,1] -0.5431798067 -0.5030271366
#> tau_sqr[1,1] 0.0105299024 0.0133094915
#> tau_sqr[2,1] 0.0063295081 0.0085237600
#> tau_sqr[3,1] -0.0011371131 0.0006865941
#> tau_sqr[4,1] -0.0006076129 0.0011599410
#> tau_sqr[5,1] -0.0056817380 -0.0002859651
#> tau_sqr[6,1] -0.0018863554 0.0030898017
#> tau_sqr[2,2] 0.0108507075 0.0137084161
#> tau_sqr[3,2] -0.0027630682 -0.0010302354
#> tau_sqr[4,2] -0.0007176545 0.0010877482
#> tau_sqr[5,2] -0.0041503193 0.0011558247
#> tau_sqr[6,2] -0.0025878975 0.0024601545
#> tau_sqr[3,3] 0.0075404163 0.0099163515
#> tau_sqr[4,3] 0.0030087052 0.0047385706
#> tau_sqr[5,3] -0.0028344455 0.0021131608
#> tau_sqr[6,3] -0.0016721519 0.0029603742
#> tau_sqr[4,4] 0.0078238625 0.0101713125
#> tau_sqr[5,4] -0.0005393147 0.0043642152
#> tau_sqr[6,4] -0.0032446621 0.0015081308
#> tau_sqr[5,5] 0.0987021940 0.1194834140
#> tau_sqr[6,5] -0.0608125308 -0.0452849853
#> tau_sqr[6,6] 0.0889923589 0.1073190175
#> i_sqr[1,1] 0.9105544323 0.9279025475
#> i_sqr[2,1] 0.9226172935 0.9376746041
#> i_sqr[3,1] 0.9436899915 0.9537497562
#> i_sqr[4,1] 0.9481545360 0.9573181364
#> i_sqr[5,1] 0.9668722384 0.9724615803
#> i_sqr[6,1] 0.9770192166 0.9808086090
confint(random, level = 0.99, lb = TRUE)
#> 0.5 % 99.5 %
#> alpha[1,1] 0.6967592331 0.7179053767
#> alpha[2,1] -0.1356671105 -0.1145475261
#> alpha[3,1] -0.1771401981 -0.1572724794
#> alpha[4,1] 0.6658987091 0.6858444150
#> alpha[5,1] 0.5083176645 0.5645453781
#> alpha[6,1] -0.5495192826 -0.4966990323
#> tau_sqr[1,1] 0.0101494559 0.0138086150
#> tau_sqr[2,1] 0.0060199534 0.0089142695
#> tau_sqr[3,1] -0.0014069587 0.0009966421
#> tau_sqr[4,1] -0.0008906037 0.0014467521
#> tau_sqr[5,1] -0.0066071682 0.0005602158
#> tau_sqr[6,1] -0.0026762032 0.0038871249
#> tau_sqr[2,2] 0.0104616214 0.0142294412
#> tau_sqr[3,2] -0.0030442262 -0.0007608539
#> tau_sqr[4,2] -0.0009834441 0.0013947324
#> tau_sqr[5,2] -0.0050025820 0.0020126698
#> tau_sqr[6,2] -0.0033933098 0.0032648898
#> tau_sqr[3,3] 0.0072210135 0.0103476678
#> tau_sqr[4,3] 0.0027504309 0.0050371473
#> tau_sqr[5,3] -0.0036366382 0.0029165021
#> tau_sqr[6,3] -0.0024094230 0.0037004267
#> tau_sqr[4,4] 0.0075070239 0.0106006504
#> tau_sqr[5,4] -0.0013661579 0.0051884873
#> tau_sqr[6,4] -0.0039818316 0.0022600588
#> tau_sqr[5,5] 0.0958650693 0.1231689344
#> tau_sqr[6,5] -0.0635429465 -0.0430900285
#> tau_sqr[6,6] 0.0864744900 0.1105899995
#> i_sqr[1,1] 0.9075017945 0.9303523990
#> i_sqr[2,1] 0.9199877081 0.9397937397
#> i_sqr[3,1] 0.9419652883 0.9551868670
#> i_sqr[4,1] 0.9466014004 0.9586042184
#> i_sqr[5,1] 0.9659271794 0.9732642969
#> i_sqr[6,1] 0.9763813488 0.9813613596- The fixed part of the random-effects model gives pooled means .
- The random part yields between-person covariances () quantifying heterogeneity in dynamics () and means () across individuals.
means <- extract(random, what = "alpha")
means
#> [1] 0.7074791 -0.1249839 -0.1670980 0.6760014 0.5364769 -0.5231016
covariances <- extract(random, what = "tau_sqr")
covariances
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 0.0118302732 7.372986e-03 -0.0002491922 0.0002701961 -0.0029657054
#> [2,] 0.0073729863 1.219111e-02 -0.0018841791 0.0001576230 -0.0014775371
#> [3,] -0.0002491922 -1.884179e-03 0.0086515593 0.0038447736 -0.0003522598
#> [4,] 0.0002701961 1.576230e-04 0.0038447736 0.0089229728 0.0018862723
#> [5,] -0.0029657054 -1.477537e-03 -0.0003522598 0.0018862723 0.1084523811
#> [6,] 0.0005965541 -6.340972e-05 0.0006454168 -0.0008607016 -0.0526615437
#> [,6]
#> [1,] 5.965541e-04
#> [2,] -6.340972e-05
#> [3,] 6.454168e-04
#> [4,] -8.607016e-04
#> [5,] -5.266154e-02
#> [6,] 9.759642e-02Finally, we compare the meta-analytic population estimates to the known generating values.
beta_pop_mean
#> [1] 0.6794619 -0.1445575 -0.1946482 0.6367942
beta_pop_cov
#> [,1] [,2] [,3] [,4]
#> [1,] 1.668405e-02 0.0099325907 0.0003451457 1.152419e-05
#> [2,] 9.932591e-03 0.0147698179 -0.0006384960 8.495547e-04
#> [3,] 3.451457e-04 -0.0006384960 0.0099156509 5.231480e-03
#> [4,] 1.152419e-05 0.0008495547 0.0052314800 1.323880e-02
nu_mu
#> [1] 0.5 -0.5
nu_sigma
#> [,1] [,2]
#> [1,] 0.10 -0.05
#> [2,] -0.05 0.10Summary
This vignette demonstrates a two-stage hierarchical estimation
approach for dynamic systems: 1. individual-level DT-VAR estimation with
stabilized measurement error, and
2. population-level meta-analysis of person-specific dynamics and
means.