Notes:
- Values in the figures represent the proportion of robust cases.
- The blue bar represents proportions greater than or equal to .90.
- The red bar represents proportions less than .90.
Type I Error
type1 <- Tree(
results_no_adj,
type = "type1",
dichotomize = TRUE
)
type1
#>
#> Conditional inference tree with 26 terminal nodes
#>
#> Response: y
#> Inputs: tauprime, n, alphabeta, mechanism, proportion, method
#> Number of observations: 8957
#>
#> 1) alphabeta == {.00(.00,.00)}; criterion = 1, statistic = 4822.126
#> 2)* weights = 1278
#> 1) alphabeta == {.00(.00,.38), .00(.00,.60), .00(.00,.71), .00(.38,.00), .00(.60,.00), .00(.71,.00)}
#> 3) method == {NBBC.COMPLETE, NBBC.FIML}; criterion = 1, statistic = 1109.392
#> 4) n <= 100; criterion = 1, statistic = 315.76
#> 5) alphabeta == {.00(.00,.71), .00(.71,.00)}; criterion = 1, statistic = 51.458
#> 6) n <= 75; criterion = 1, statistic = 108.376
#> 7) n <= 50; criterion = 1, statistic = 25.615
#> 8)* weights = 56
#> 7) n > 50
#> 9)* weights = 56
#> 6) n > 75
#> 10)* weights = 56
#> 5) alphabeta == {.00(.00,.38), .00(.00,.60), .00(.38,.00), .00(.60,.00)}
#> 11)* weights = 336
#> 4) n > 100
#> 12) alphabeta == {.00(.00,.60), .00(.00,.71), .00(.60,.00), .00(.71,.00)}; criterion = 1, statistic = 256.035
#> 13)* weights = 560
#> 12) alphabeta == {.00(.00,.38), .00(.38,.00)}
#> 14) n <= 200; criterion = 1, statistic = 113.879
#> 15)* weights = 112
#> 14) n > 200
#> 16)* weights = 168
#> 3) method == {MC.COMPLETE, MC.FIML, MC.MI, NBPC.COMPLETE, NBPC.FIML, SIG.COMPLETE, SIG.FIML, SIG.MI}
#> 17) n <= 50; criterion = 1, statistic = 237.481
#> 18) alphabeta == {.00(.00,.71), .00(.60,.00), .00(.71,.00)}; criterion = 1, statistic = 154.161
#> 19) method == {MC.COMPLETE, NBPC.COMPLETE, NBPC.FIML, SIG.COMPLETE}; criterion = 1, statistic = 162.331
#> 20)* weights = 108
#> 19) method == {MC.FIML, MC.MI, SIG.FIML, SIG.MI}
#> 21) alphabeta == {.00(.60,.00)}; criterion = 1, statistic = 43.72
#> 22)* weights = 96
#> 21) alphabeta == {.00(.00,.71), .00(.71,.00)}
#> 23) method == {MC.FIML, SIG.FIML}; criterion = 1, statistic = 42.169
#> 24)* weights = 96
#> 23) method == {MC.MI, SIG.MI}
#> 25)* weights = 96
#> 18) alphabeta == {.00(.00,.38), .00(.00,.60), .00(.38,.00)}
#> 26) method == {MC.COMPLETE, MC.FIML, MC.MI, NBPC.COMPLETE, SIG.COMPLETE, SIG.FIML, SIG.MI}; criterion = 1, statistic = 57.616
#> 27) tauprime <= 0.1414214; criterion = 1, statistic = 37.307
#> 28) alphabeta == {.00(.00,.60)}; criterion = 1, statistic = 43.7
#> 29) method == {MC.FIML, SIG.FIML}; criterion = 1, statistic = 41.075
#> 30)* weights = 24
#> 29) method == {MC.COMPLETE, MC.MI, NBPC.COMPLETE, SIG.COMPLETE, SIG.MI}
#> 31)* weights = 30
#> 28) alphabeta == {.00(.00,.38), .00(.38,.00)}
#> 32) method == {MC.MI}; criterion = 1, statistic = 46.813
#> 33) alphabeta == {.00(.00,.38)}; criterion = 1, statistic = 23
#> 34)* weights = 12
#> 33) alphabeta == {.00(.38,.00)}
#> 35)* weights = 12
#> 32) method == {MC.COMPLETE, MC.FIML, NBPC.COMPLETE, SIG.COMPLETE, SIG.FIML, SIG.MI}
#> 36)* weights = 84
#> 27) tauprime > 0.1414214
#> 37)* weights = 162
#> 26) method == {NBPC.FIML}
#> 38) alphabeta == {.00(.00,.38), .00(.38,.00)}; criterion = 1, statistic = 47.333
#> 39)* weights = 48
#> 38) alphabeta == {.00(.00,.60)}
#> 40)* weights = 24
#> 17) n > 50
#> 41) alphabeta == {.00(.00,.38), .00(.00,.60), .00(.00,.71), .00(.38,.00)}; criterion = 1, statistic = 87.193
#> 42)* weights = 3695
#> 41) alphabeta == {.00(.60,.00), .00(.71,.00)}
#> 43) tauprime <= 0; criterion = 1, statistic = 29.366
#> 44) alphabeta == {.00(.60,.00)}; criterion = 1, statistic = 25.26
#> 45)* weights = 231
#> 44) alphabeta == {.00(.71,.00)}
#> 46)* weights = 231
#> 43) tauprime > 0
#> 47) method == {MC.FIML, SIG.FIML}; criterion = 1, statistic = 37.46
#> 48) alphabeta == {.00(.71,.00)}; criterion = 1, statistic = 18.63
#> 49)* weights = 252
#> 48) alphabeta == {.00(.60,.00)}
#> 50)* weights = 252
#> 47) method == {MC.COMPLETE, MC.MI, NBPC.COMPLETE, NBPC.FIML, SIG.COMPLETE, SIG.MI}
#> 51)* weights = 882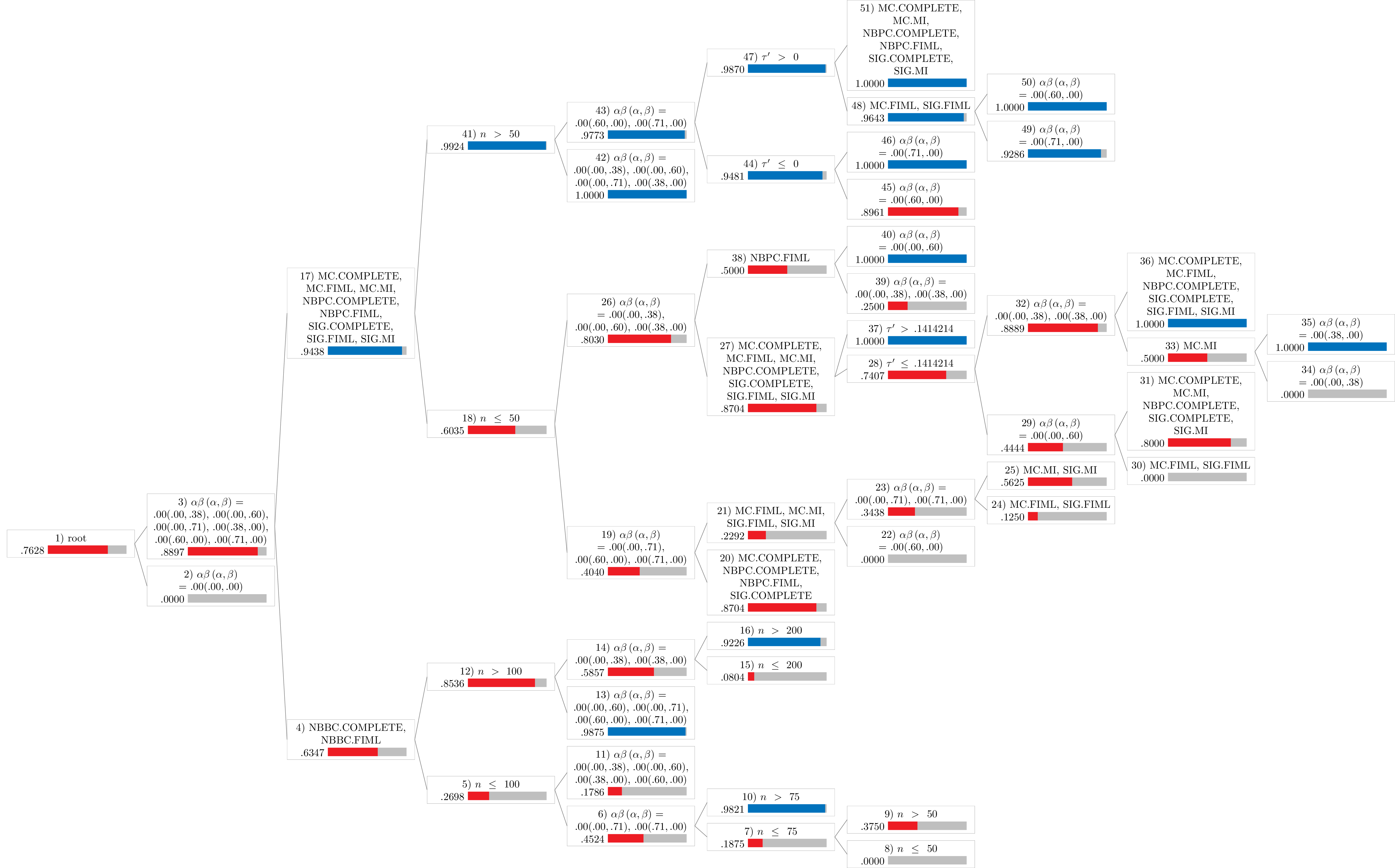
Type I Error Rate (Dichotomized)
Statistical Power
power <- Tree(
results_no_adj,
type = "power",
dichotomize = TRUE
)
power
#>
#> Conditional inference tree with 26 terminal nodes
#>
#> Response: y
#> Inputs: tauprime, n, alphabeta, mechanism, proportion, method
#> Number of observations: 9915
#>
#> 1) alphabeta == {.14(.38,.38), .23(.60,.38), .27(.71,.38)}; criterion = 1, statistic = 1046.041
#> 2) n <= 75; criterion = 1, statistic = 651.181
#> 3) tauprime <= 0.3605551; criterion = 1, statistic = 268.144
#> 4) mechanism == {MAR, MCAR}; criterion = 1, statistic = 75.478
#> 5) alphabeta == {.14(.38,.38), .27(.71,.38)}; criterion = 1, statistic = 62.816
#> 6)* weights = 432
#> 5) alphabeta == {.23(.60,.38)}
#> 7) tauprime <= 0.1414214; criterion = 1, statistic = 58.848
#> 8)* weights = 144
#> 7) tauprime > 0.1414214
#> 9) n <= 50; criterion = 1, statistic = 50.714
#> 10)* weights = 36
#> 9) n > 50
#> 11)* weights = 36
#> 4) mechanism == {COMPLETE}
#> 12) n <= 50; criterion = 1, statistic = 33.327
#> 13)* weights = 36
#> 12) n > 50
#> 14)* weights = 36
#> 3) tauprime > 0.3605551
#> 15) n <= 50; criterion = 1, statistic = 85.332
#> 16) alphabeta == {.14(.38,.38), .27(.71,.38)}; criterion = 1, statistic = 79.13
#> 17) mechanism == {COMPLETE}; criterion = 1, statistic = 37.421
#> 18)* weights = 8
#> 17) mechanism == {MAR, MCAR}
#> 19)* weights = 72
#> 16) alphabeta == {.23(.60,.38)}
#> 20) method == {MC.FIML, MC.MI, NBBC.FIML, SIG.FIML, SIG.MI}; criterion = 1, statistic = 39
#> 21)* weights = 30
#> 20) method == {MC.COMPLETE, NBBC.COMPLETE, NBPC.COMPLETE, NBPC.FIML, SIG.COMPLETE}
#> 22)* weights = 10
#> 15) n > 50
#> 23) alphabeta == {.14(.38,.38)}; criterion = 1, statistic = 30.852
#> 24) method == {MC.MI, NBPC.FIML}; criterion = 1, statistic = 39
#> 25)* weights = 12
#> 24) method == {MC.COMPLETE, MC.FIML, NBBC.COMPLETE, NBBC.FIML, NBPC.COMPLETE, SIG.COMPLETE, SIG.FIML, SIG.MI}
#> 26)* weights = 28
#> 23) alphabeta == {.23(.60,.38), .27(.71,.38)}
#> 27)* weights = 80
#> 2) n > 75
#> 28) tauprime <= 0.1414214; criterion = 1, statistic = 142.443
#> 29) n <= 100; criterion = 1, statistic = 105.352
#> 30) alphabeta == {.27(.71,.38)}; criterion = 1, statistic = 102.82
#> 31) mechanism == {COMPLETE}; criterion = 1, statistic = 37.421
#> 32)* weights = 8
#> 31) mechanism == {MAR, MCAR}
#> 33)* weights = 72
#> 30) alphabeta == {.14(.38,.38), .23(.60,.38)}
#> 34) method == {NBPC.FIML}; criterion = 1, statistic = 59
#> 35)* weights = 24
#> 34) method == {MC.COMPLETE, MC.FIML, MC.MI, NBBC.COMPLETE, NBBC.FIML, NBPC.COMPLETE, SIG.COMPLETE, SIG.FIML, SIG.MI}
#> 36) alphabeta == {.23(.60,.38)}; criterion = 1, statistic = 38.208
#> 37) tauprime <= 0; criterion = 1, statistic = 52.895
#> 38)* weights = 34
#> 37) tauprime > 0
#> 39)* weights = 34
#> 36) alphabeta == {.14(.38,.38)}
#> 40)* weights = 68
#> 29) n > 100
#> 41)* weights = 1200
#> 28) tauprime > 0.1414214
#> 42)* weights = 1440
#> 1) alphabeta == {.23(.38,.60), .27(.38,.71), .36(.60,.60), .43(.60,.71), .43(.71,.60), .51(.71,.71)}
#> 43) alphabeta == {.23(.38,.60), .27(.38,.71)}; criterion = 1, statistic = 460.011
#> 44) n <= 50; criterion = 1, statistic = 187.5
#> 45) method == {MC.FIML, MC.MI, NBBC.FIML, NBPC.COMPLETE, NBPC.FIML, SIG.FIML, SIG.MI}; criterion = 1, statistic = 246.644
#> 46) mechanism == {COMPLETE}; criterion = 1, statistic = 71.994
#> 47)* weights = 7
#> 46) mechanism == {MAR, MCAR}
#> 48)* weights = 251
#> 45) method == {MC.COMPLETE, NBBC.COMPLETE, SIG.COMPLETE}
#> 49)* weights = 21
#> 44) n > 50
#> 50)* weights = 1959
#> 43) alphabeta == {.36(.60,.60), .43(.60,.71), .43(.71,.60), .51(.71,.71)}
#> 51)* weights = 3837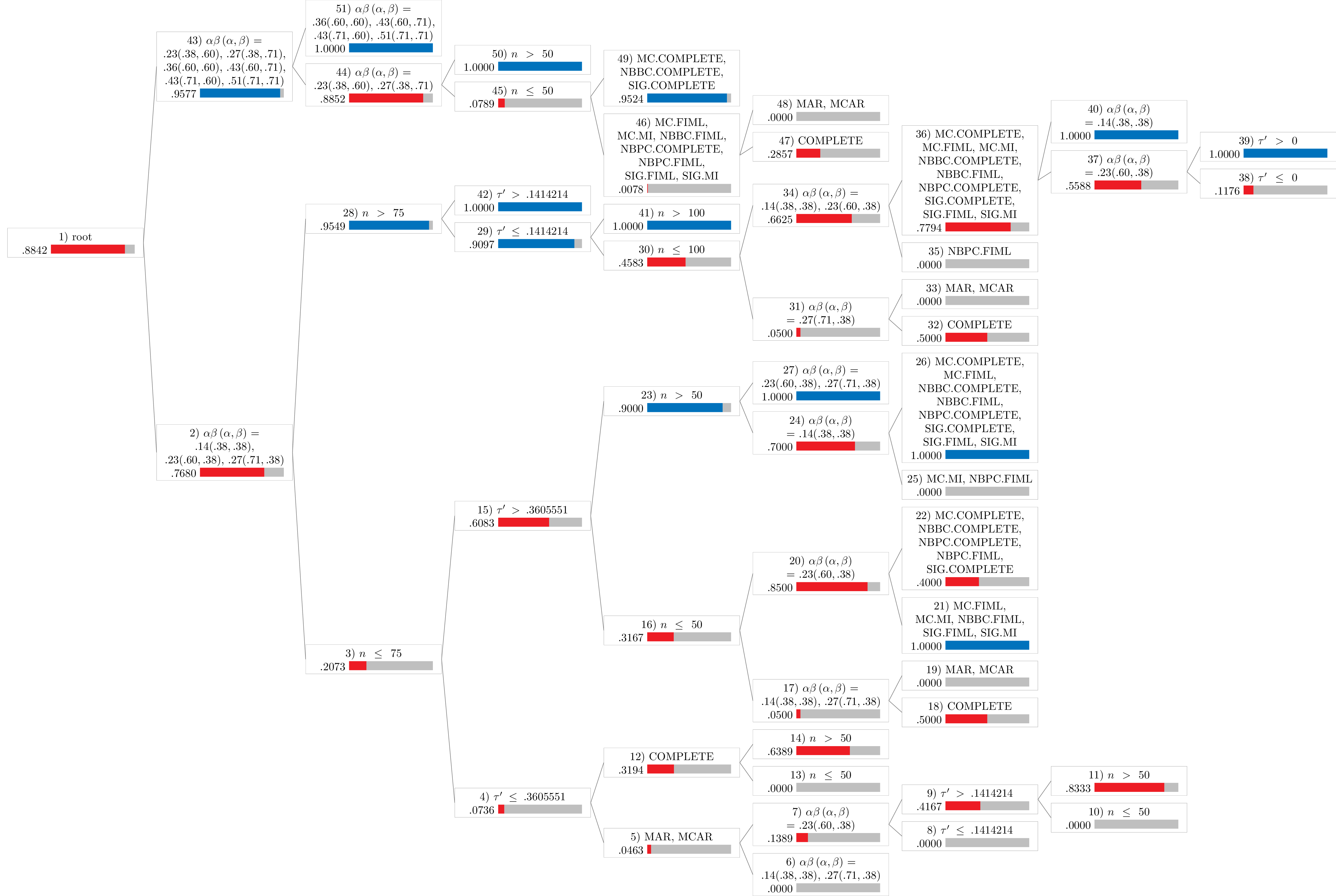
Statistical Power (Dichotomized)
Miss Rate
miss <- Tree(
results_no_adj,
type = "miss",
dichotomize = TRUE
)
miss
#>
#> Conditional inference tree with 12 terminal nodes
#>
#> Response: y
#> Inputs: tauprime, n, alphabeta, mechanism, proportion, method
#> Number of observations: 6691
#>
#> 1) n <= 50; criterion = 1, statistic = 70.723
#> 2) tauprime <= 0.1414214; criterion = 1, statistic = 66.756
#> 3) method == {MC.COMPLETE, MC.FIML, NBBC.COMPLETE, NBPC.COMPLETE}; criterion = 1, statistic = 35.415
#> 4)* weights = 162
#> 3) method == {MC.MI, NBBC.FIML, NBPC.FIML}
#> 5)* weights = 321
#> 2) tauprime > 0.1414214
#> 6) alphabeta == {.14(.38,.38), .23(.38,.60), .23(.60,.38), .27(.38,.71), .27(.71,.38), .36(.60,.60), .43(.71,.60)}; criterion = 1, statistic = 69.745
#> 7) tauprime <= 0.3605551; criterion = 1, statistic = 30.072
#> 8) method == {MC.FIML}; criterion = 1, statistic = 44.61
#> 9) alphabeta == {.14(.38,.38), .23(.38,.60), .27(.38,.71), .36(.60,.60), .43(.71,.60)}; criterion = 1, statistic = 41
#> 10)* weights = 30
#> 9) alphabeta == {.23(.60,.38), .27(.71,.38)}
#> 11)* weights = 12
#> 8) method == {MC.COMPLETE, MC.MI, NBBC.COMPLETE, NBBC.FIML, NBPC.COMPLETE, NBPC.FIML}
#> 12)* weights = 147
#> 7) tauprime > 0.3605551
#> 13) alphabeta == {.14(.38,.38), .23(.38,.60), .36(.60,.60)}; criterion = 1, statistic = 37.365
#> 14)* weights = 81
#> 13) alphabeta == {.23(.60,.38), .27(.71,.38)}
#> 15)* weights = 54
#> 6) alphabeta == {.43(.60,.71)}
#> 16)* weights = 27
#> 1) n > 50
#> 17) alphabeta == {.14(.38,.38), .23(.38,.60), .23(.60,.38), .27(.71,.38), .36(.60,.60), .43(.60,.71), .43(.71,.60), .51(.71,.71)}; criterion = 1, statistic = 144.263
#> 18) alphabeta == {.14(.38,.38), .23(.38,.60), .23(.60,.38), .36(.60,.60), .43(.60,.71), .43(.71,.60), .51(.71,.71)}; criterion = 1, statistic = 36.018
#> 19)* weights = 4534
#> 18) alphabeta == {.27(.71,.38)}
#> 20)* weights = 756
#> 17) alphabeta == {.27(.38,.71)}
#> 21) tauprime <= 0.1414214; criterion = 1, statistic = 35.119
#> 22)* weights = 378
#> 21) tauprime > 0.1414214
#> 23)* weights = 189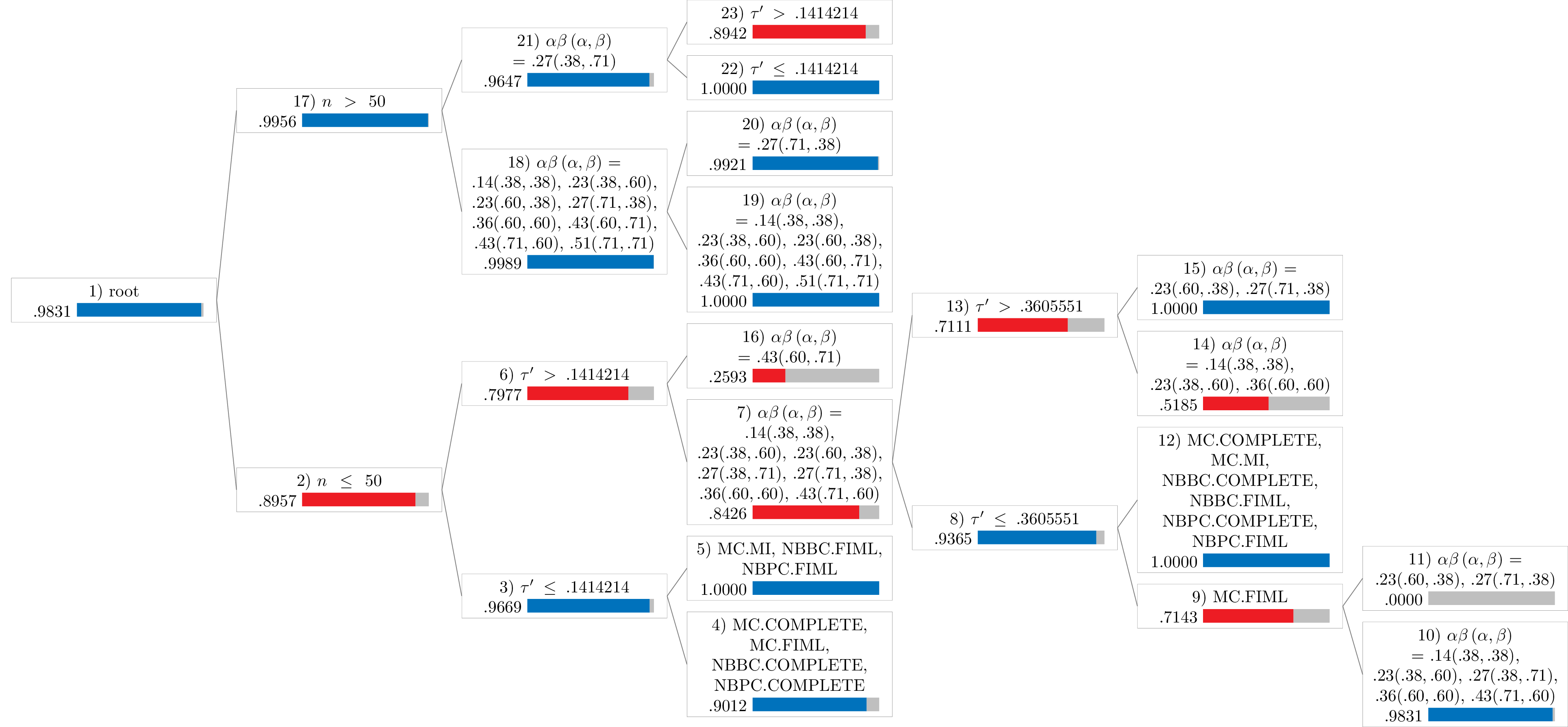
Miss Rate (Dichotomized)