Proportion of Missing Values
Ivan Jacob Agaloos Pesigan
Source:vignettes/proportion-missing.Rmd
proportion-missing.RmdThe following presents more details on the proportion of missing values used in the simulation study.
Amputation
prop <- 0.30
data_missing <- AmputeData(
data_complete,
mech = "MCAR",
prop = prop
)Missing Data Patterns
The matrix below shows the missing data patterns in the amputed data.
mice::md.pattern(data_missing)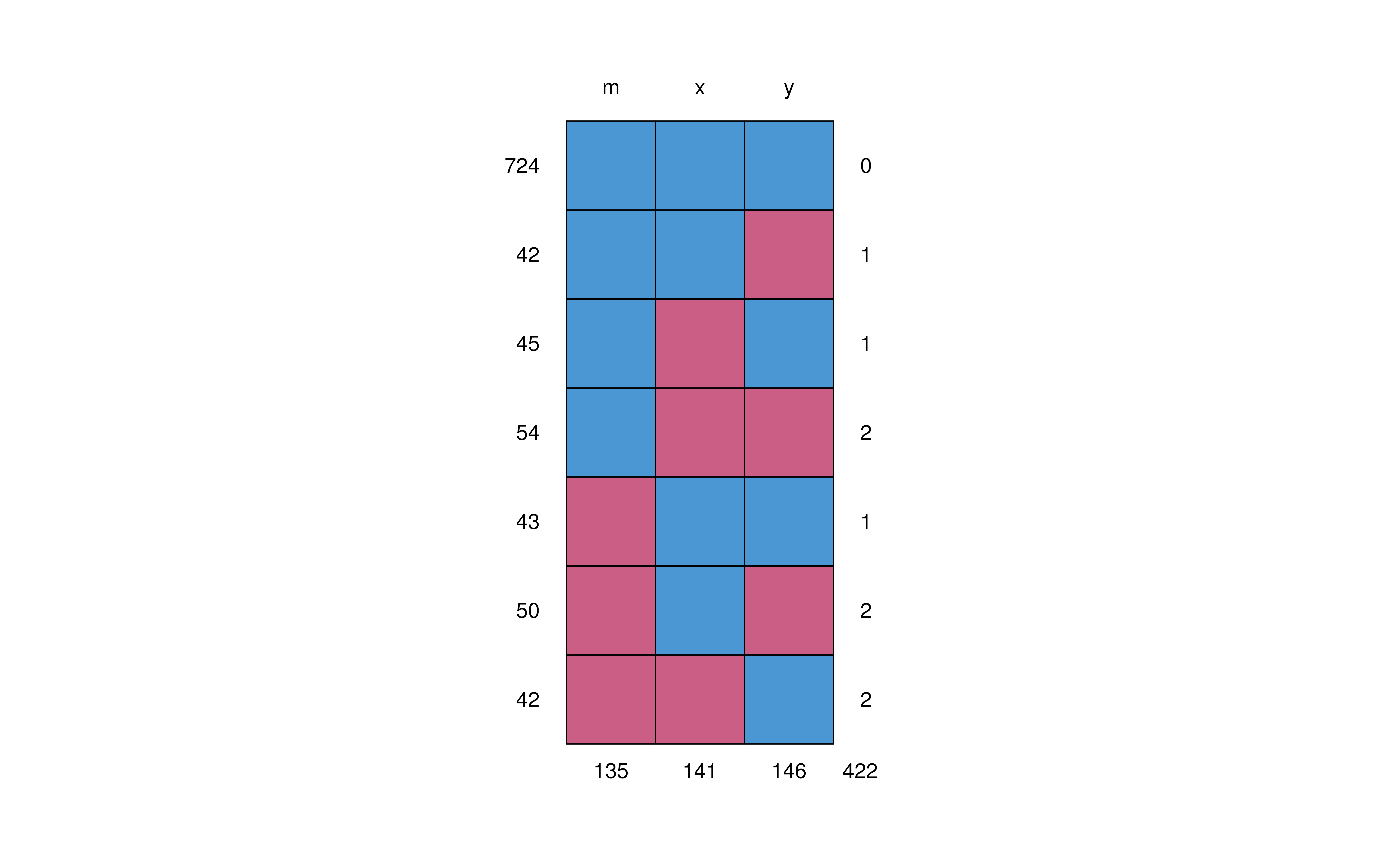
#> m x y
#> 724 1 1 1 0
#> 42 1 1 0 1
#> 45 1 0 1 1
#> 54 1 0 0 2
#> 43 0 1 1 1
#> 50 0 1 0 2
#> 42 0 0 1 2
#> 135 141 146 422Proportion of missing values by row
This is the number of rows with missing values divided by the total
number of rows. This is the value of the prop argument in
AmputeData() which is a wrapper around the
mice::ampute() function.
sum_row <- sum(!complete.cases(data_missing))
prop_row <- sum_row / n
c(
sum_row = sum_row,
prop_row = prop_row
)
#> sum_row prop_row
#> 276.000 0.276Proportion of missing values by column
This is the number of rows per column with missing values divided by the total number of rows.
sum_col <- colSums(is.na(data_missing))
names(sum_col) <- paste0(
"sum_col_",
names(sum_col)
)
prop_col <- sum_col / n
names(prop_col) <- gsub("sum", "prop", names(sum_col))
c(
sum_col,
prop_col
)
#> sum_col_x sum_col_m sum_col_y prop_col_x prop_col_m prop_col_y
#> 141.000 135.000 146.000 0.141 0.135 0.146Empirical Proportion of Missing Values
Below is a simple simulation to show the empirical proportions for
the combination of sample size n and prop used
in the study for 5000 replications.
MAR
| n | prop | sum_row | prop_row | sum_col_x | sum_col_m | sum_col_y | prop_col_x | prop_col_m | prop_col_y | sum_cell | prop_cell | reps |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 0.1 | 4.91 | 0.1 | 2.47 | 2.46 | 2.45 | 0.05 | 0.05 | 0.05 | 7.37 | 0.05 | 5000 |
| 75 | 0.1 | 7.43 | 0.1 | 3.71 | 3.73 | 3.70 | 0.05 | 0.05 | 0.05 | 11.14 | 0.05 | 5000 |
| 100 | 0.1 | 10.04 | 0.1 | 5.01 | 5.01 | 5.04 | 0.05 | 0.05 | 0.05 | 15.06 | 0.05 | 5000 |
| 150 | 0.1 | 15.02 | 0.1 | 7.48 | 7.48 | 7.54 | 0.05 | 0.05 | 0.05 | 22.51 | 0.05 | 5000 |
| 200 | 0.1 | 19.81 | 0.1 | 9.89 | 9.87 | 9.90 | 0.05 | 0.05 | 0.05 | 29.66 | 0.05 | 5000 |
| 250 | 0.1 | 24.91 | 0.1 | 12.45 | 12.46 | 12.42 | 0.05 | 0.05 | 0.05 | 37.33 | 0.05 | 5000 |
| 500 | 0.1 | 50.00 | 0.1 | 25.04 | 24.94 | 25.06 | 0.05 | 0.05 | 0.05 | 75.03 | 0.05 | 5000 |
| 1000 | 0.1 | 100.13 | 0.1 | 50.21 | 49.96 | 49.97 | 0.05 | 0.05 | 0.05 | 150.15 | 0.05 | 5000 |
| 50 | 0.2 | 9.87 | 0.2 | 4.94 | 4.95 | 4.92 | 0.10 | 0.10 | 0.10 | 14.81 | 0.10 | 5000 |
| 75 | 0.2 | 14.90 | 0.2 | 7.43 | 7.46 | 7.47 | 0.10 | 0.10 | 0.10 | 22.37 | 0.10 | 5000 |
| 100 | 0.2 | 19.91 | 0.2 | 9.95 | 9.96 | 9.99 | 0.10 | 0.10 | 0.10 | 29.89 | 0.10 | 5000 |
| 150 | 0.2 | 30.05 | 0.2 | 15.04 | 15.03 | 15.06 | 0.10 | 0.10 | 0.10 | 45.13 | 0.10 | 5000 |
| 200 | 0.2 | 39.84 | 0.2 | 19.88 | 19.89 | 19.96 | 0.10 | 0.10 | 0.10 | 59.72 | 0.10 | 5000 |
| 250 | 0.2 | 49.88 | 0.2 | 25.06 | 24.89 | 24.83 | 0.10 | 0.10 | 0.10 | 74.78 | 0.10 | 5000 |
| 500 | 0.2 | 100.03 | 0.2 | 49.95 | 50.07 | 50.00 | 0.10 | 0.10 | 0.10 | 150.01 | 0.10 | 5000 |
| 1000 | 0.2 | 200.17 | 0.2 | 100.40 | 99.96 | 99.89 | 0.10 | 0.10 | 0.10 | 300.26 | 0.10 | 5000 |
| 50 | 0.3 | 14.85 | 0.3 | 7.42 | 7.39 | 7.43 | 0.15 | 0.15 | 0.15 | 22.24 | 0.15 | 5000 |
| 75 | 0.3 | 22.35 | 0.3 | 11.17 | 11.16 | 11.23 | 0.15 | 0.15 | 0.15 | 33.56 | 0.15 | 5000 |
| 100 | 0.3 | 29.86 | 0.3 | 14.90 | 14.95 | 14.95 | 0.15 | 0.15 | 0.15 | 44.81 | 0.15 | 5000 |
| 150 | 0.3 | 44.88 | 0.3 | 22.40 | 22.53 | 22.48 | 0.15 | 0.15 | 0.15 | 67.41 | 0.15 | 5000 |
| 200 | 0.3 | 60.06 | 0.3 | 30.04 | 29.98 | 30.08 | 0.15 | 0.15 | 0.15 | 90.10 | 0.15 | 5000 |
| 250 | 0.3 | 74.75 | 0.3 | 37.53 | 37.33 | 37.25 | 0.15 | 0.15 | 0.15 | 112.11 | 0.15 | 5000 |
| 500 | 0.3 | 149.97 | 0.3 | 74.87 | 75.20 | 74.93 | 0.15 | 0.15 | 0.15 | 224.99 | 0.15 | 5000 |
| 1000 | 0.3 | 299.77 | 0.3 | 150.08 | 149.76 | 149.83 | 0.15 | 0.15 | 0.15 | 449.68 | 0.15 | 5000 |
MCAR
| n | prop | sum_row | prop_row | sum_col_x | sum_col_m | sum_col_y | prop_col_x | prop_col_m | prop_col_y | sum_cell | prop_cell | reps |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 0.1 | 4.99 | 0.1 | 2.49 | 2.49 | 2.50 | 0.05 | 0.05 | 0.05 | 7.48 | 0.05 | 5000 |
| 75 | 0.1 | 7.47 | 0.1 | 3.74 | 3.74 | 3.71 | 0.05 | 0.05 | 0.05 | 11.20 | 0.05 | 5000 |
| 100 | 0.1 | 10.01 | 0.1 | 4.99 | 4.99 | 5.02 | 0.05 | 0.05 | 0.05 | 15.00 | 0.05 | 5000 |
| 150 | 0.1 | 15.05 | 0.1 | 7.51 | 7.52 | 7.52 | 0.05 | 0.05 | 0.05 | 22.56 | 0.05 | 5000 |
| 200 | 0.1 | 20.03 | 0.1 | 10.11 | 10.02 | 9.94 | 0.05 | 0.05 | 0.05 | 30.08 | 0.05 | 5000 |
| 250 | 0.1 | 25.04 | 0.1 | 12.52 | 12.51 | 12.54 | 0.05 | 0.05 | 0.05 | 37.57 | 0.05 | 5000 |
| 500 | 0.1 | 50.11 | 0.1 | 25.01 | 25.15 | 24.99 | 0.05 | 0.05 | 0.05 | 75.15 | 0.05 | 5000 |
| 1000 | 0.1 | 100.08 | 0.1 | 50.13 | 49.99 | 50.02 | 0.05 | 0.05 | 0.05 | 150.14 | 0.05 | 5000 |
| 50 | 0.2 | 9.99 | 0.2 | 5.03 | 4.97 | 5.00 | 0.10 | 0.10 | 0.10 | 15.00 | 0.10 | 5000 |
| 75 | 0.2 | 14.94 | 0.2 | 7.49 | 7.45 | 7.44 | 0.10 | 0.10 | 0.10 | 22.38 | 0.10 | 5000 |
| 100 | 0.2 | 19.99 | 0.2 | 9.97 | 9.97 | 10.01 | 0.10 | 0.10 | 0.10 | 29.95 | 0.10 | 5000 |
| 150 | 0.2 | 30.04 | 0.2 | 15.05 | 14.98 | 15.00 | 0.10 | 0.10 | 0.10 | 45.03 | 0.10 | 5000 |
| 200 | 0.2 | 40.04 | 0.2 | 20.09 | 20.04 | 19.97 | 0.10 | 0.10 | 0.10 | 60.10 | 0.10 | 5000 |
| 250 | 0.2 | 50.05 | 0.2 | 25.08 | 25.04 | 25.02 | 0.10 | 0.10 | 0.10 | 75.14 | 0.10 | 5000 |
| 500 | 0.2 | 100.09 | 0.2 | 49.98 | 50.13 | 49.99 | 0.10 | 0.10 | 0.10 | 150.11 | 0.10 | 5000 |
| 1000 | 0.2 | 200.09 | 0.2 | 100.06 | 100.02 | 99.93 | 0.10 | 0.10 | 0.10 | 300.02 | 0.10 | 5000 |
| 50 | 0.3 | 15.02 | 0.3 | 7.52 | 7.50 | 7.49 | 0.15 | 0.15 | 0.15 | 22.51 | 0.15 | 5000 |
| 75 | 0.3 | 22.47 | 0.3 | 11.27 | 11.22 | 11.19 | 0.15 | 0.15 | 0.15 | 33.67 | 0.15 | 5000 |
| 100 | 0.3 | 29.99 | 0.3 | 15.00 | 14.96 | 14.97 | 0.15 | 0.15 | 0.15 | 44.93 | 0.15 | 5000 |
| 150 | 0.3 | 45.05 | 0.3 | 22.60 | 22.46 | 22.47 | 0.15 | 0.15 | 0.15 | 67.54 | 0.15 | 5000 |
| 200 | 0.3 | 60.11 | 0.3 | 30.15 | 30.03 | 30.01 | 0.15 | 0.15 | 0.15 | 90.20 | 0.15 | 5000 |
| 250 | 0.3 | 75.06 | 0.3 | 37.61 | 37.51 | 37.50 | 0.15 | 0.15 | 0.15 | 112.62 | 0.15 | 5000 |
| 500 | 0.3 | 150.10 | 0.3 | 75.06 | 75.05 | 74.99 | 0.15 | 0.15 | 0.15 | 225.10 | 0.15 | 5000 |
| 1000 | 0.3 | 300.14 | 0.3 | 150.09 | 150.00 | 150.00 | 0.15 | 0.15 | 0.15 | 450.09 | 0.15 | 5000 |
Code to perform the simulation.
bar <- function(mech) {
foo <- function(x,
reps = 5000,
mech,
seed = 42) {
set.seed(42)
n <- x[1]
prop <- x[2]
data_complete <- GenData(n = n)
colMeans(
do.call(
what = "rbind",
args = lapply(
X = 1:reps,
FUN = function(i, mech) {
data_missing <- AmputeData(
data_complete,
mech = mech,
prop = prop
)
sum_row <- sum(!complete.cases(data_missing))
prop_row <- sum_row / n
row <- c(
sum_row = sum_row,
prop_row = prop_row
)
sum_col <- colSums(is.na(data_missing))
names(sum_col) <- paste0(
"sum_col_",
names(sum_col)
)
prop_col <- sum_col / n
names(prop_col) <- gsub("sum", "prop", names(sum_col))
col <- c(
sum_col,
prop_col
)
sum_cell <- sum(is.na(data_missing))
prop_cell <- sum_cell / (n * 3)
prop_cell
cell <- c(
sum_cell = sum_cell,
prop_cell = prop_cell
)
c(
n = n,
prop = prop,
row,
col,
cell,
reps = reps
)
},
mech = mech
)
)
)
}
do.call(
what = "rbind",
args = lapply(
X = as.data.frame(
t(
expand.grid(
n = unique(manMCMedMiss::params$n),
prop = 1:3 * .10
)
)
),
FUN = foo,
mech = mech
)
)
}
prop_mar <- bar("MAR")
prop_mcar <- bar("MCAR")