Illustration 3 - Regions of Significance
Ivan Jacob Agaloos Pesigan
Source:vignettes/fig-example-3.Rmd
fig-example-3.RmdThis vignette accompanies Illustration 3. The goal of the
illustration is to visualize regions of significance, that is, a range
of time intervals, where the effects are significantly different from
zero, using confidence intervals for the direct, indirect, and total
effects from the continuous-time vector autoregressive model drift
matrix
and process noise covariance matrix
for a range of time intervals. This example features plot
methods for the DeltaMed, MCMed,
BootMed, DeltaMedStd, MCMedStd,
and BootMedStd functions from the cTMed
package.
Continuous-Time Vector Autoregressive Model Estimates
The object fit contains the fitted dynr
model. Data is generated using the
manCTMed::IllustrationGenData function. The model was
fitted using the manCTMed::IllustrationFitDynr.
summary(fit)
#> Coefficients:
#> Estimate Std. Error t value ci.lower ci.upper Pr(>|t|)
#> phi_11 -0.1157754 0.0174197 -6.646 -0.1499173 -0.0816334 <2e-16 ***
#> phi_12 0.0203747 0.0550571 0.370 -0.0875352 0.1282846 0.3557
#> phi_13 -0.0070992 0.0535302 -0.133 -0.1120165 0.0978181 0.4472
#> phi_21 -0.1226909 0.0187426 -6.546 -0.1594258 -0.0859560 <2e-16 ***
#> phi_22 -0.8819998 0.0653717 -13.492 -1.0101259 -0.7538737 <2e-16 ***
#> phi_23 0.4737224 0.0594135 7.973 0.3572740 0.5901708 <2e-16 ***
#> phi_31 -0.0877980 0.0178492 -4.919 -0.1227817 -0.0528143 <2e-16 ***
#> phi_32 0.0595814 0.0586861 1.015 -0.0554414 0.1746041 0.1550
#> phi_33 -0.6636176 0.0616495 -10.764 -0.7844485 -0.5427867 <2e-16 ***
#> sigma_11 0.0954899 0.0049239 19.393 0.0858393 0.1051405 <2e-16 ***
#> sigma_12 0.0009624 0.0040987 0.235 -0.0070710 0.0089957 0.4072
#> sigma_13 0.0070297 0.0040069 1.754 -0.0008237 0.0148830 0.0397 *
#> sigma_22 0.1027182 0.0078961 13.009 0.0872422 0.1181942 <2e-16 ***
#> sigma_23 0.0009240 0.0049248 0.188 -0.0087285 0.0105764 0.4256
#> sigma_33 0.0991956 0.0074695 13.280 0.0845558 0.1138355 <2e-16 ***
#> theta_11 0.1016133 0.0014901 68.194 0.0986928 0.1045338 <2e-16 ***
#> theta_22 0.0996380 0.0015668 63.592 0.0965671 0.1027089 <2e-16 ***
#> theta_33 0.0992561 0.0015460 64.202 0.0962260 0.1022862 <2e-16 ***
#> mu0_1 -0.1217526 0.0521444 -2.335 -0.2239538 -0.0195514 0.0098 **
#> mu0_2 0.0160785 0.0279075 0.576 -0.0386192 0.0707762 0.2823
#> mu0_3 0.0204163 0.0288928 0.707 -0.0362125 0.0770451 0.2399
#> sigma0_11 0.3346315 0.0446548 7.494 0.2471097 0.4221533 <2e-16 ***
#> sigma0_12 -0.0644525 0.0177503 -3.631 -0.0992424 -0.0296626 0.0001 ***
#> sigma0_13 -0.0519690 0.0179974 -2.888 -0.0872433 -0.0166946 0.0019 **
#> sigma0_22 0.0691741 0.0127707 5.417 0.0441439 0.0942043 <2e-16 ***
#> sigma0_23 0.0364469 0.0098132 3.714 0.0172134 0.0556805 0.0001 ***
#> sigma0_33 0.0792446 0.0137057 5.782 0.0523819 0.1061072 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> -2 log-likelihood value at convergence = 32662.46
#> AIC = 32716.46
#> BIC = 32919.11Parameter Estimates
We extract parameter estimates from the fit object.
# drift matrix
phi <- matrix(
data = coef(fit)[
c(
"phi_11", "phi_21", "phi_31",
"phi_12", "phi_22", "phi_32",
"phi_13", "phi_23", "phi_33"
)
],
nrow = 3
)
# column names and row names are needed to define the indirect effects
colnames(phi) <- rownames(phi) <- c(
"conflict",
"knowledge",
"competence"
)
# process noise covariance matrix
sigma <- matrix(
data = coef(fit)[
c(
"sigma_11", "sigma_12", "sigma_13",
"sigma_12", "sigma_22", "sigma_23",
"sigma_13", "sigma_23", "sigma_33"
)
],
nrow = 3
)
# measurement error covariance matrix
theta <- matrix(
data = coef(fit)[
c(
"theta_11", 0, 0,
0, "theta_22", 0,
0, 0, "theta_33"
)
],
nrow = 3
)
# initial condition
mu0 <- coef(fit)[
c(
"mu0_1", "mu0_2", "mu0_3"
)
]
sigma0 <- matrix(
data = coef(fit)[
c(
"sigma0_11", "sigma0_12", "sigma0_13",
"sigma0_12", "sigma0_22", "sigma0_23",
"sigma0_13", "sigma0_23", "sigma0_33"
)
],
nrow = 3
)Sampling Variance-Covariance Matrix of Parameter Estimates
# combining the drift matrix and the process noise covariance matrix
phi_sigma_names <- c(
"phi_11", "phi_21", "phi_31",
"phi_12", "phi_22", "phi_32",
"phi_13", "phi_23", "phi_33",
"sigma_11", "sigma_12", "sigma_13",
"sigma_22", "sigma_23",
"sigma_33"
)
vcov_theta <- vcov(fit)[phi_sigma_names, phi_sigma_names]
phi_names <- c(
"phi_11", "phi_21", "phi_31",
"phi_12", "phi_22", "phi_32",
"phi_13", "phi_23", "phi_33"
)
vcov_phi_vec <- vcov_theta[phi_names, phi_names]Delta Method Confidence Intervals For The Direct, Indirect, and Total Effects
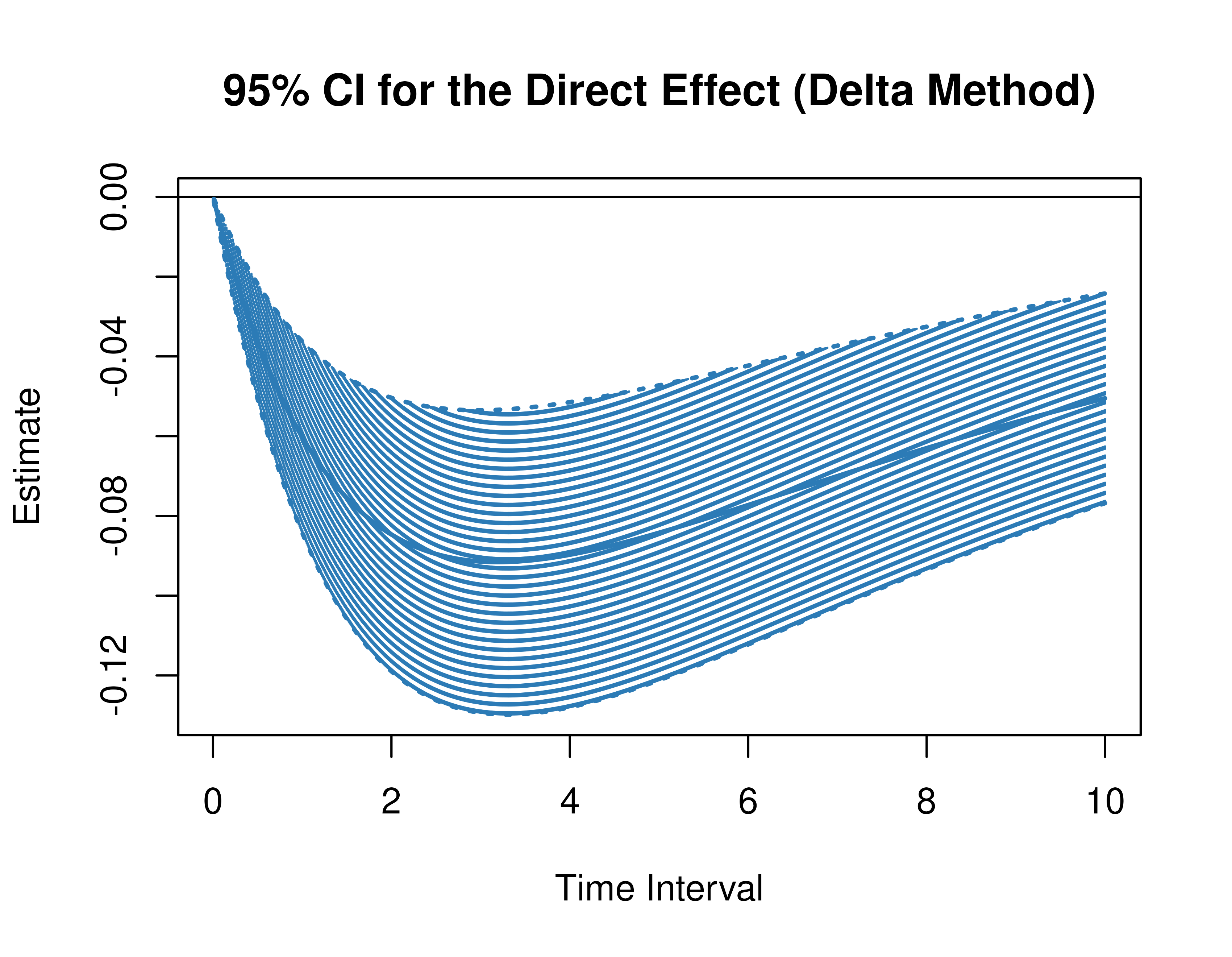
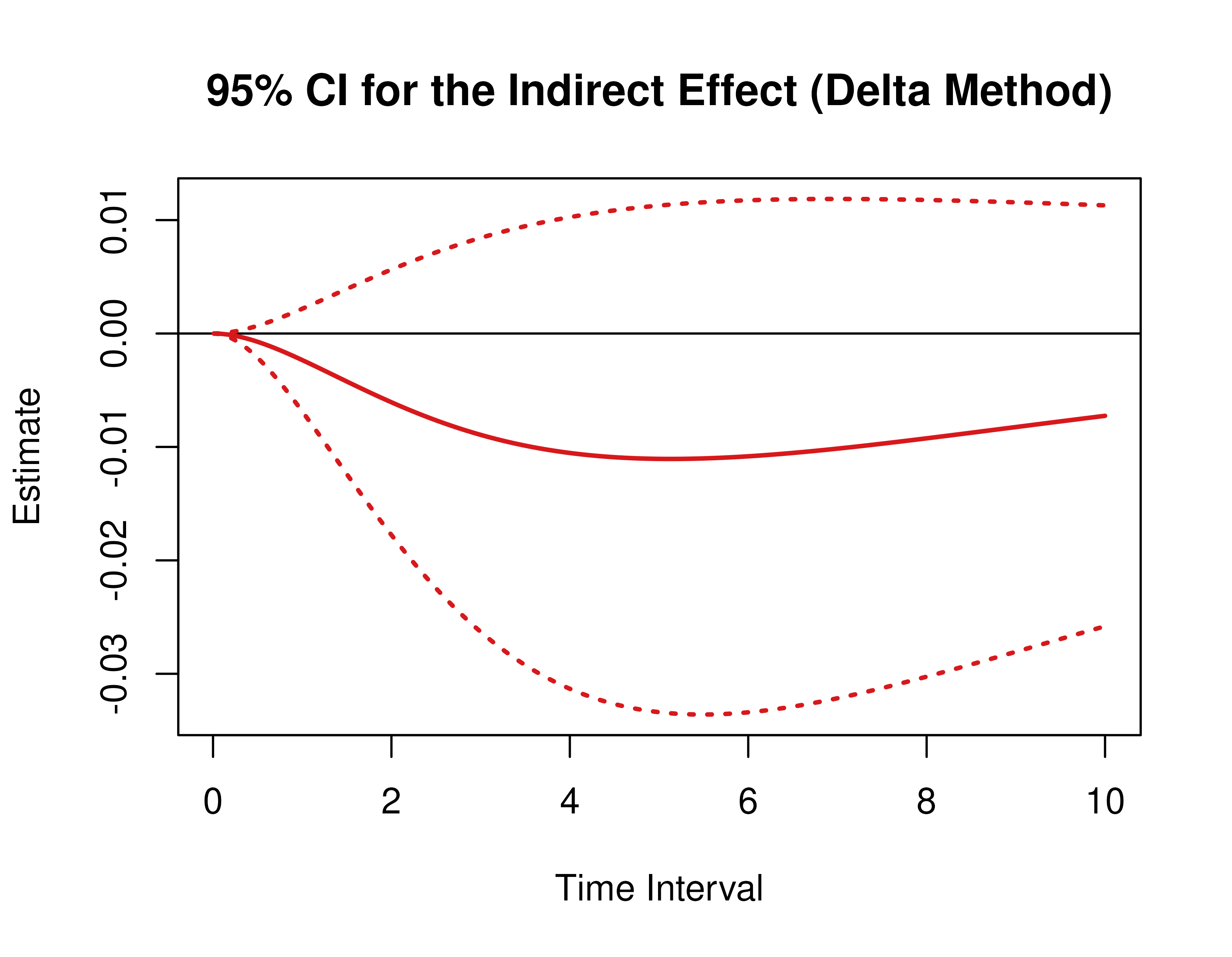
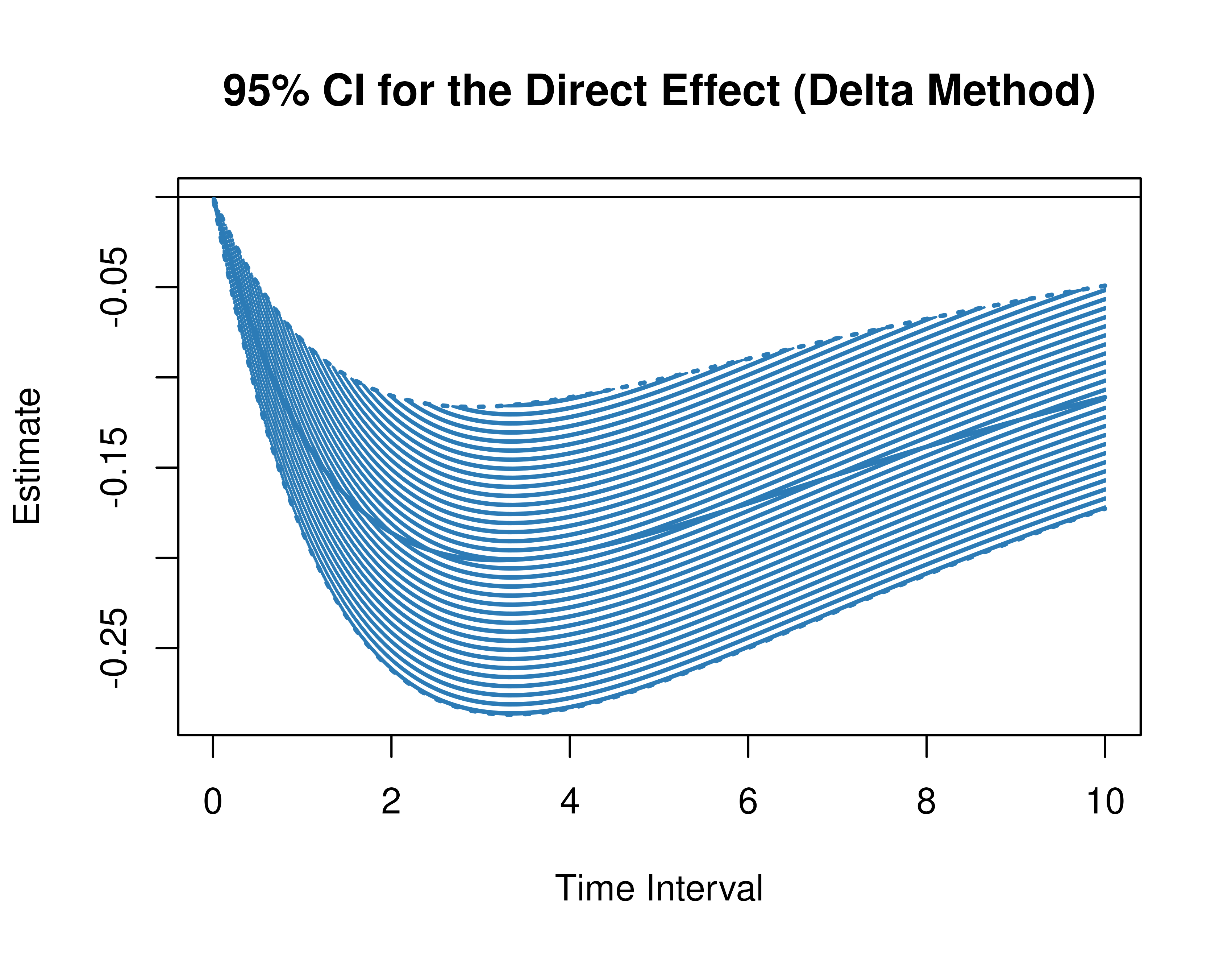
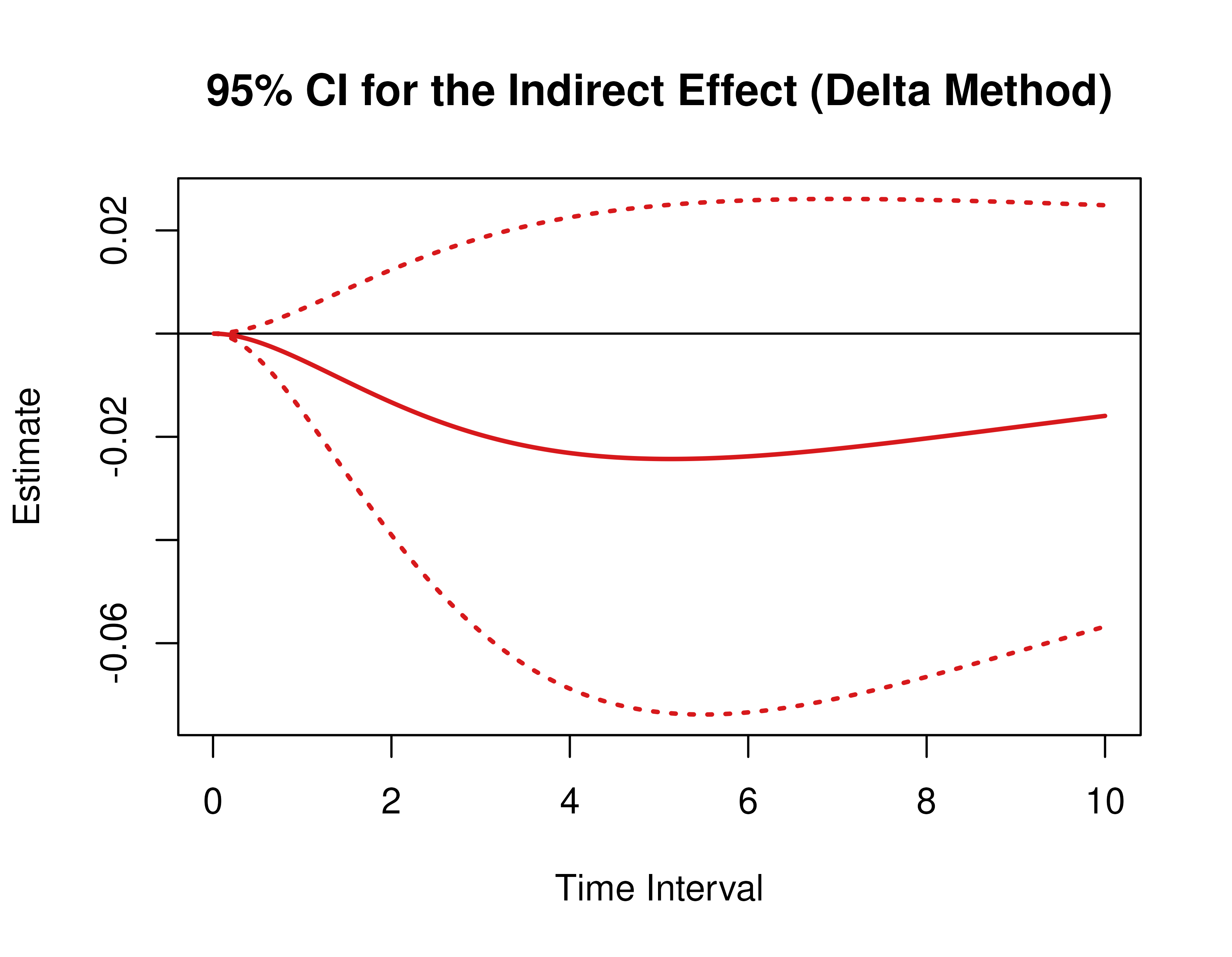
Using the DeltaMed function from the cTMed
package, confidence intervals for the direct, indirect, and total
effects for a long sequence of time interval values are generated. This
makes regions of significance more visible. Consider using the
ncores argument to use multiple cores when the
delta_t vector is long. The plot method for
the DeltaMed presents the regions of significance visually
represented by shaded areas in the plot.
delta <- DeltaMed(
phi = phi,
vcov_phi_vec = vcov_phi_vec,
delta_t = delta_t,
from = "conflict",
to = "competence",
med = "knowledge",
ncores = parallel::detectCores()
)
plot(delta)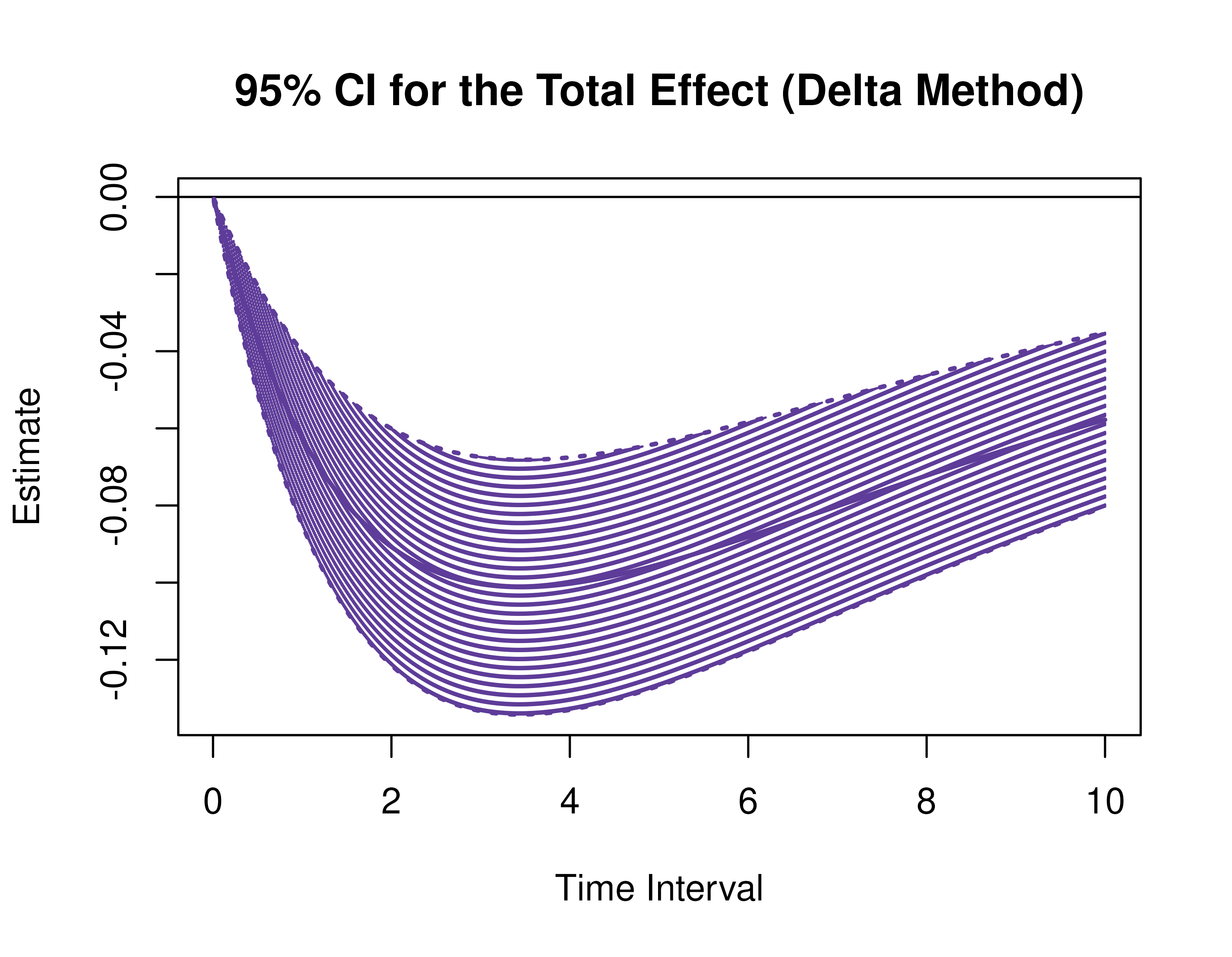
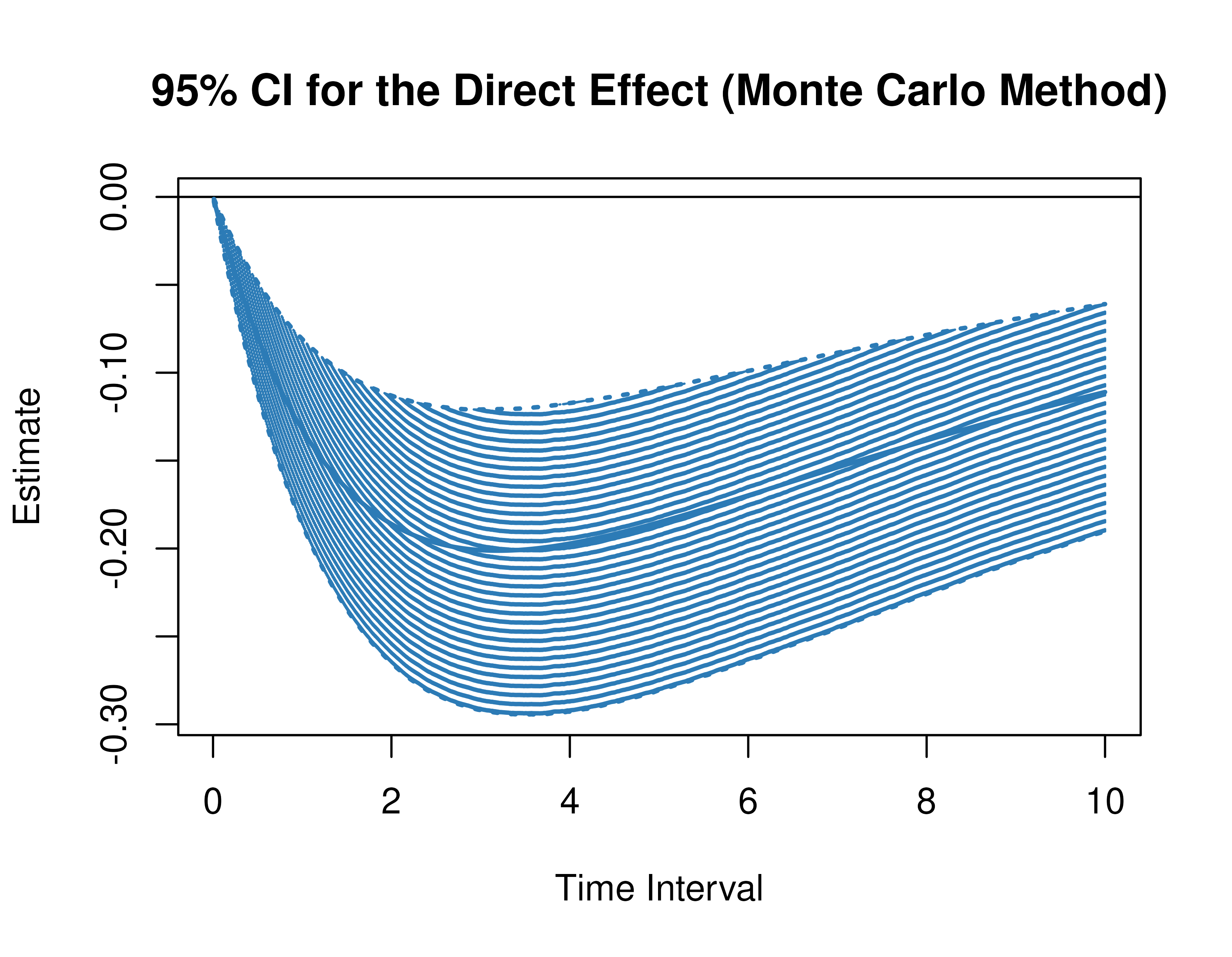
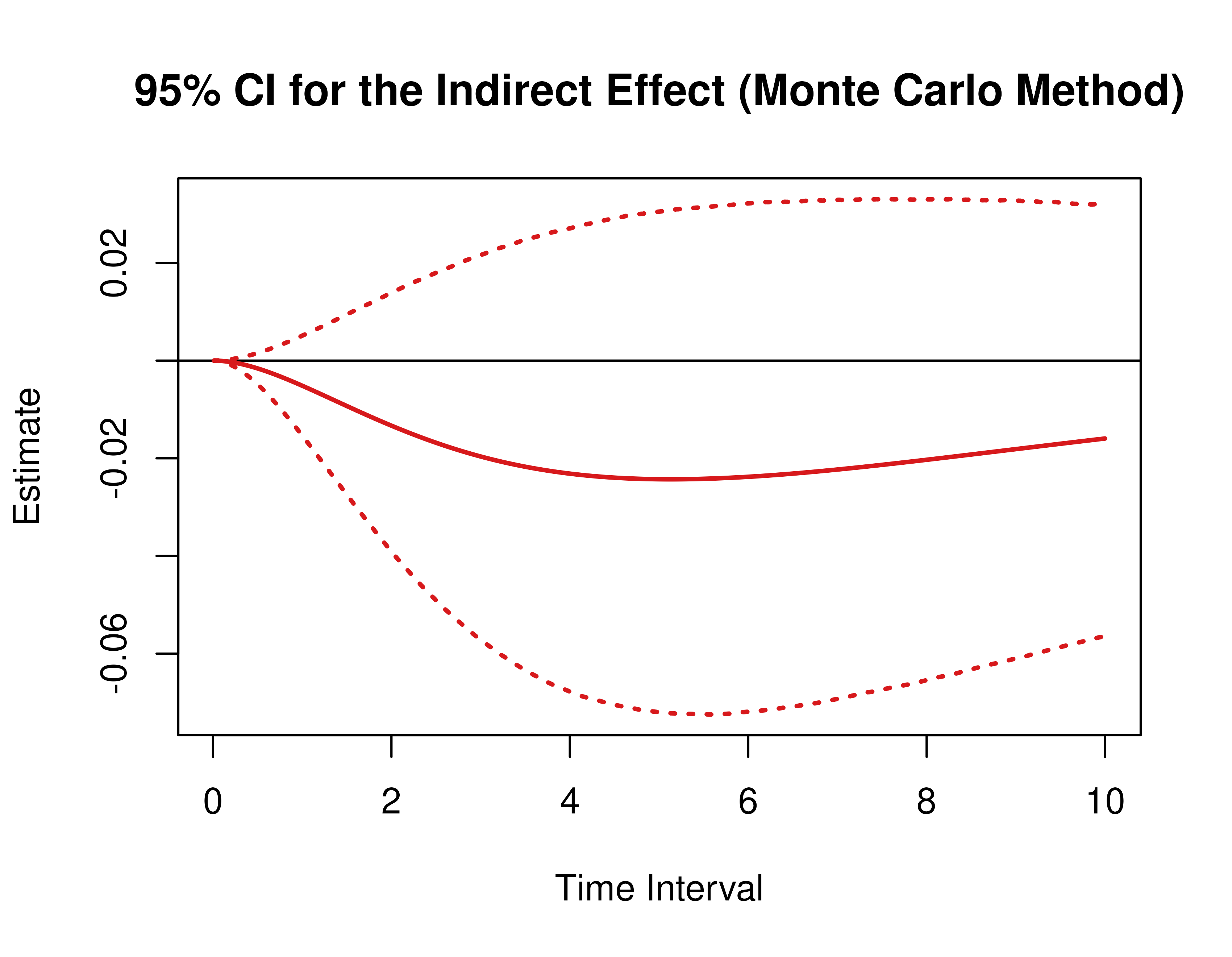
Monte Carlo Method Confidence Intervals For The Direct, Indirect, and Total Effects
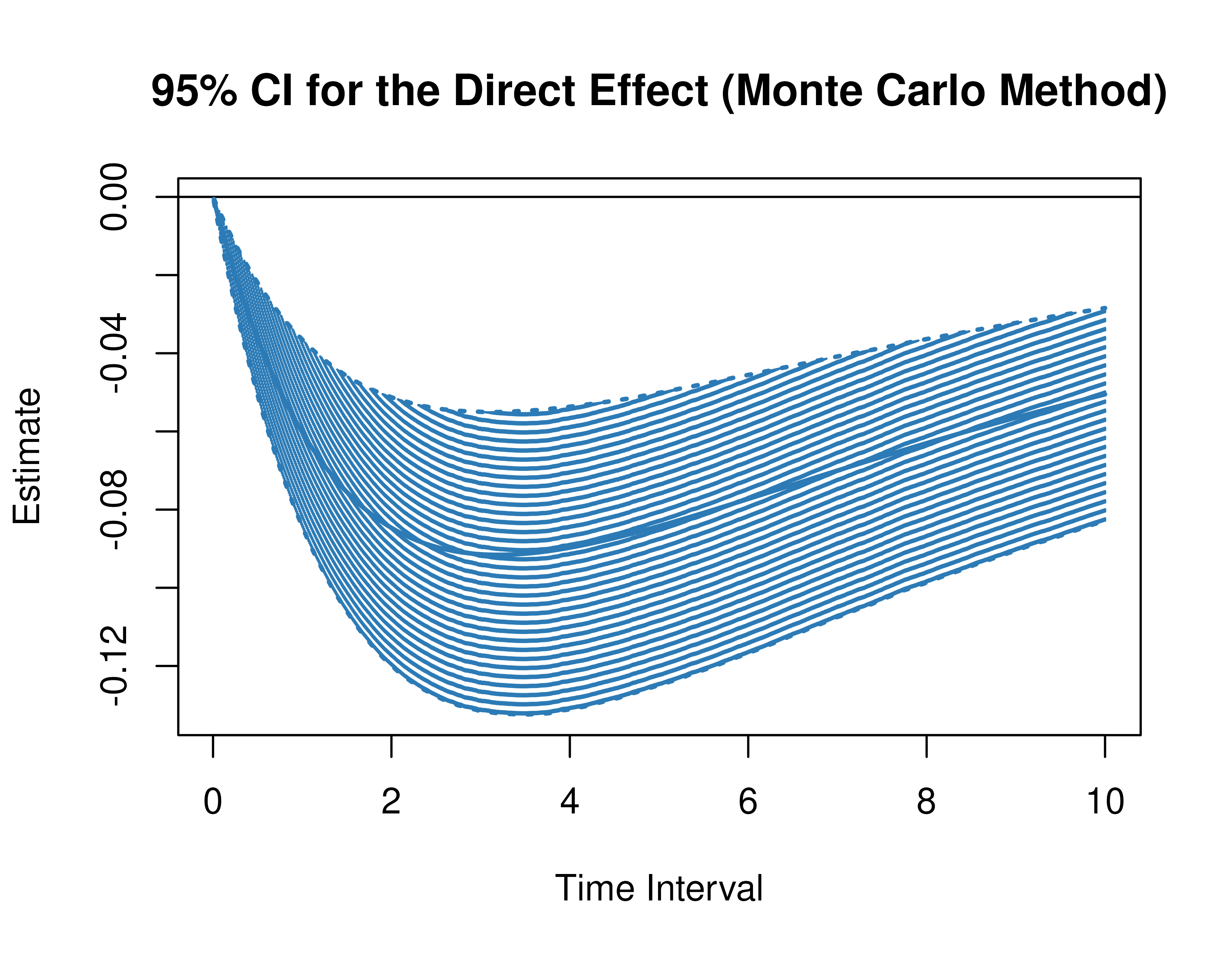
Using the MCMed function from the cTMed
package, confidence intervals for the direct, indirect, and total
effects for a long sequence of time interval values are generated. This
makes regions of significance more visible. Consider using the
ncores argument to use multiple cores when the
delta_t vector is long. The plot method for
the MCMed presents the regions of significance visually
represented by shaded areas in the plot.
mc <- MCMed(
phi = phi,
vcov_phi_vec = vcov_phi_vec,
delta_t = delta_t,
from = "conflict",
to = "competence",
med = "knowledge",
R = 20000L,
ncores = parallel::detectCores(), # use multiple cores
seed = 42
)
plot(mc)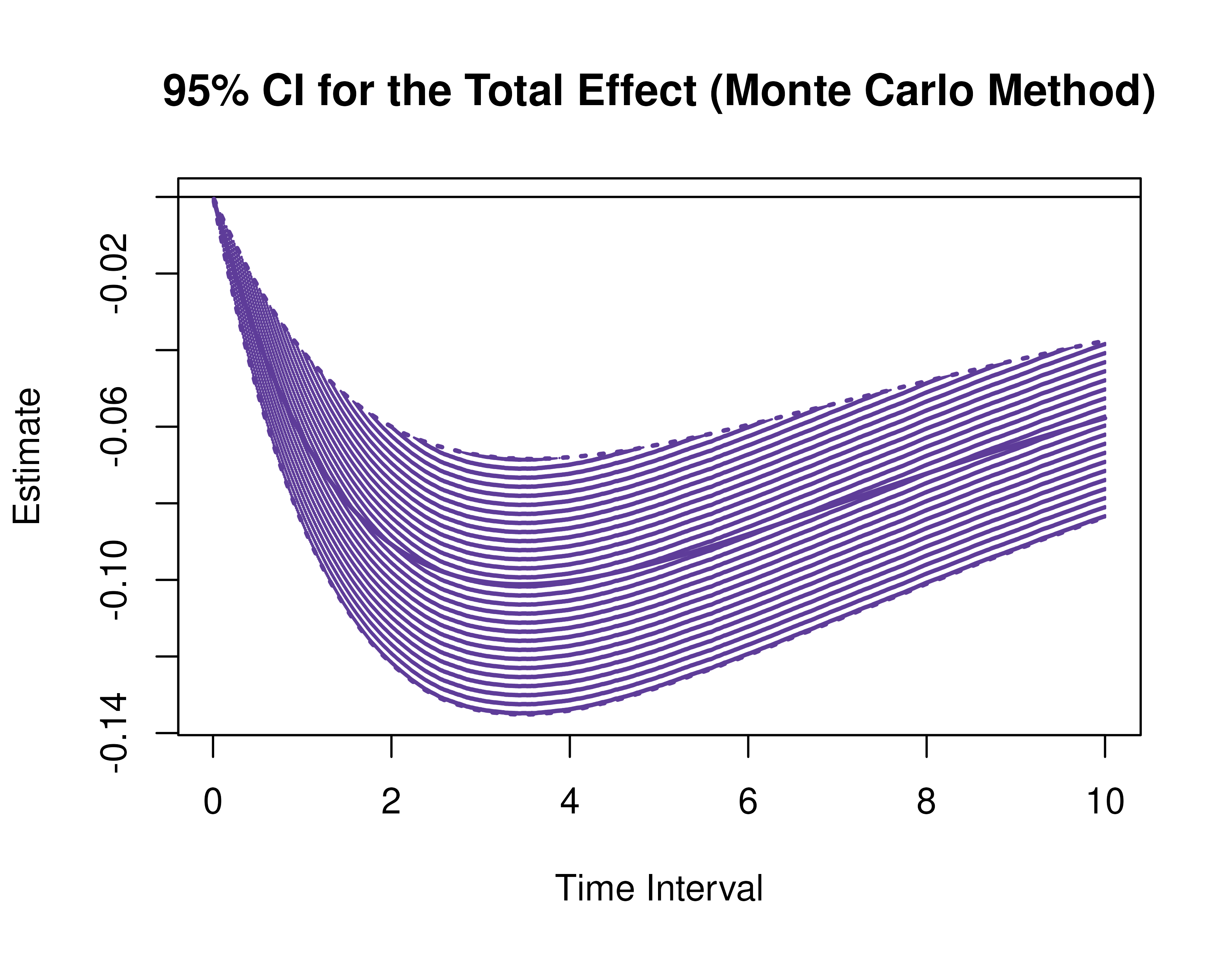

Parametric Bootstrap Method Confidence Intervals For The Direct, Indirect, and Total Effects
The parametric bootstrap approach involves generating data from the
parametric model and fitting the model on the generated data multiple
times. The data generation and model fitting is hadled by the
bootStateSpace package. The estimated parameters are passed
as arguments to the PBSSMOUFixed function from the
bootStateSpace package, which generates a parametric
bootstrap sampling distribution of the parameter estimates. The argument
R specifies the number of bootstrap replications. The
generated data and model estimates are be stored in path
using the specified prefix for the file names. The
ncores = parallel::detectCores() argument instructs the
function to use all available CPU cores in the system.
NOTE: Fitting the CT-VAR model multiple times is computationally intensive.
R <- 1000L
path <- root$find_file(
".setup",
"data-raw"
)
prefix <- "illustration_pb"
phi_hat <- phi
sigma_hat <- sigma
boot <- PBSSMOUFixed(
R = R,
path = path,
prefix = prefix,
n = 133,
time = 101,
delta_t = 0.10,
mu0 = mu0,
sigma0_l = t(chol(sigma0)),
mu = c(0, 0, 0),
phi = phi,
sigma_l = t(chol(sigma)),
nu = c(0, 0, 0),
lambda = diag(3),
theta_l = t(chol(theta)),
mu0_fixed = FALSE,
sigma0_fixed = FALSE,
ncores = parallel::detectCores(),
seed = 42,
clean = FALSE
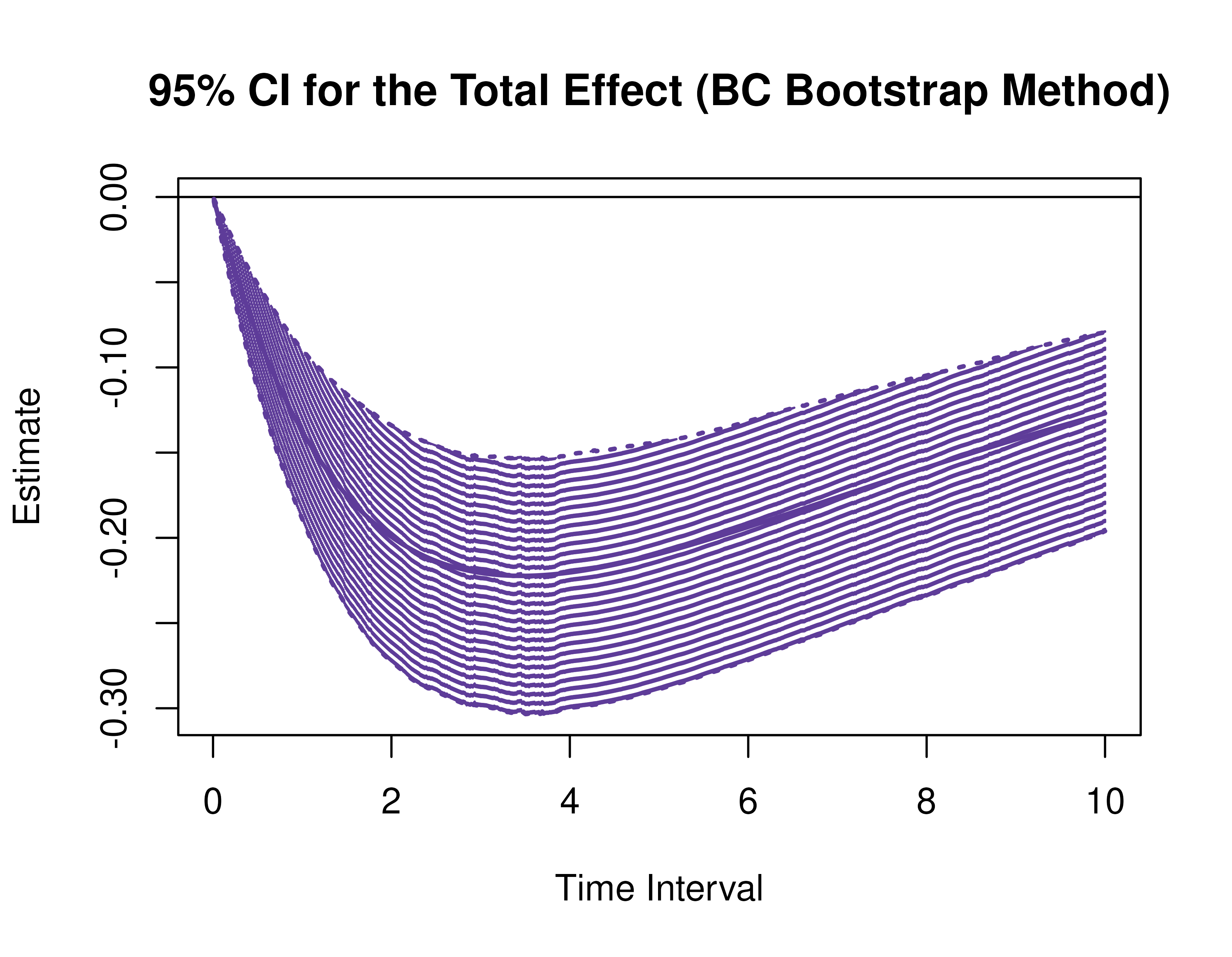
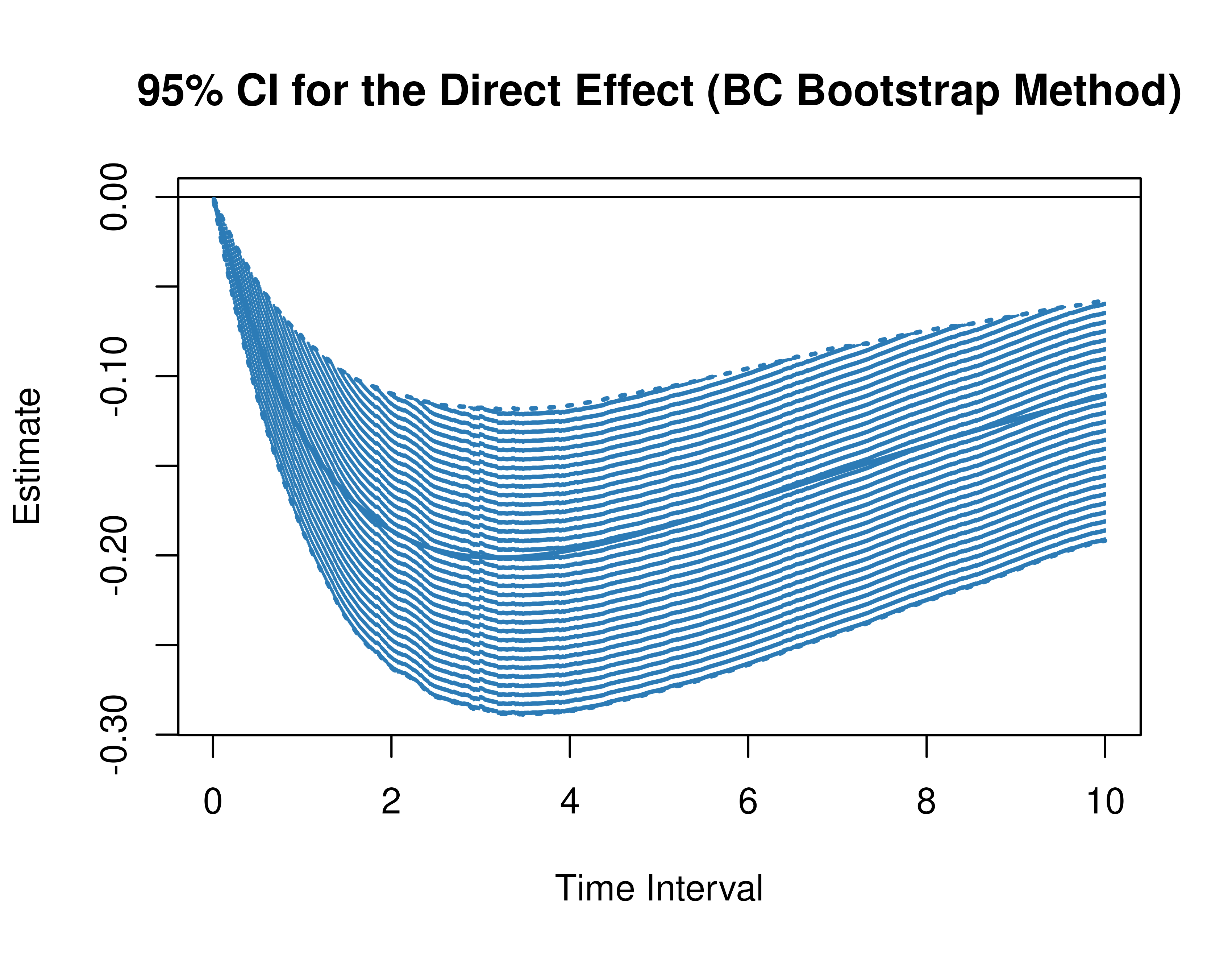
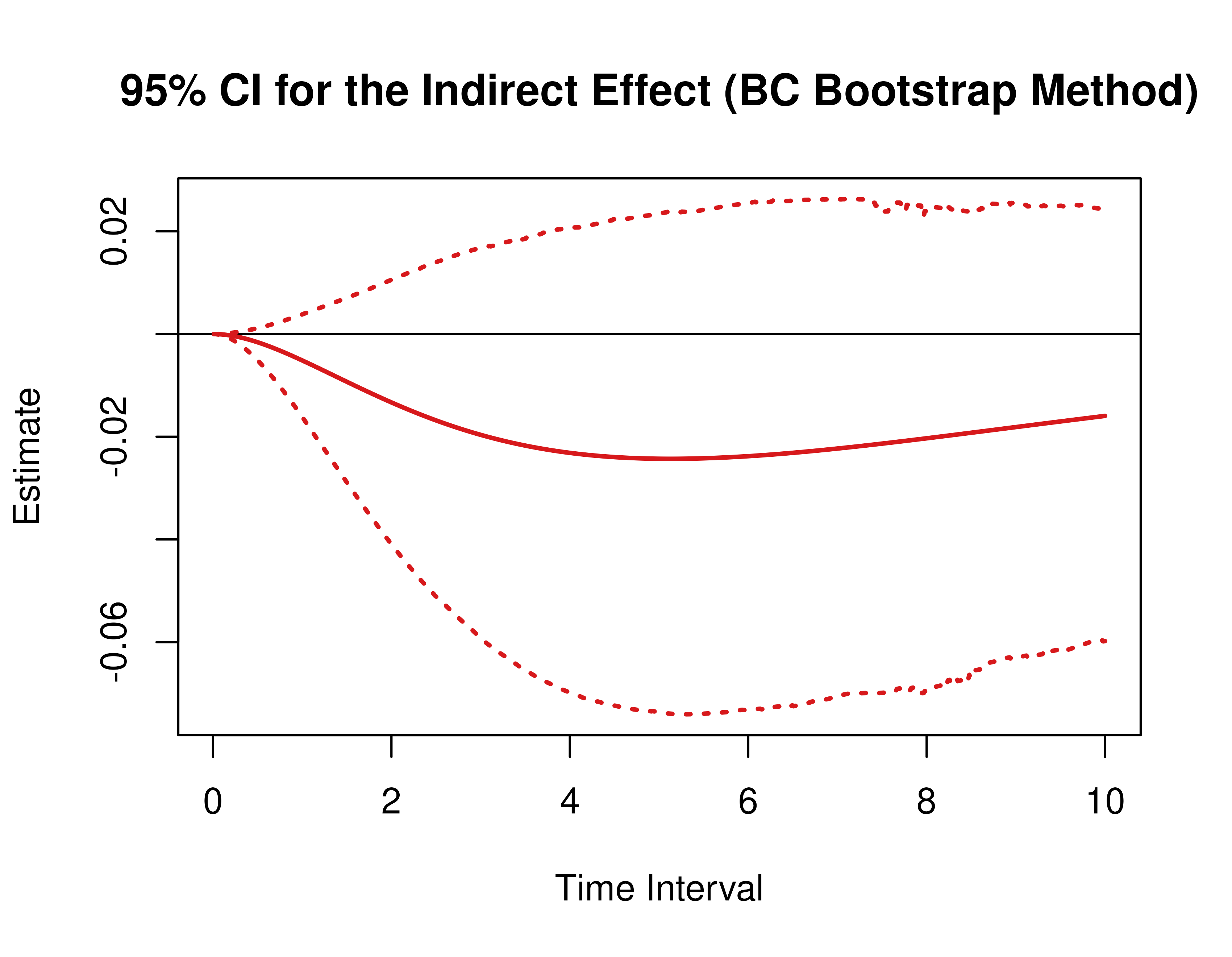
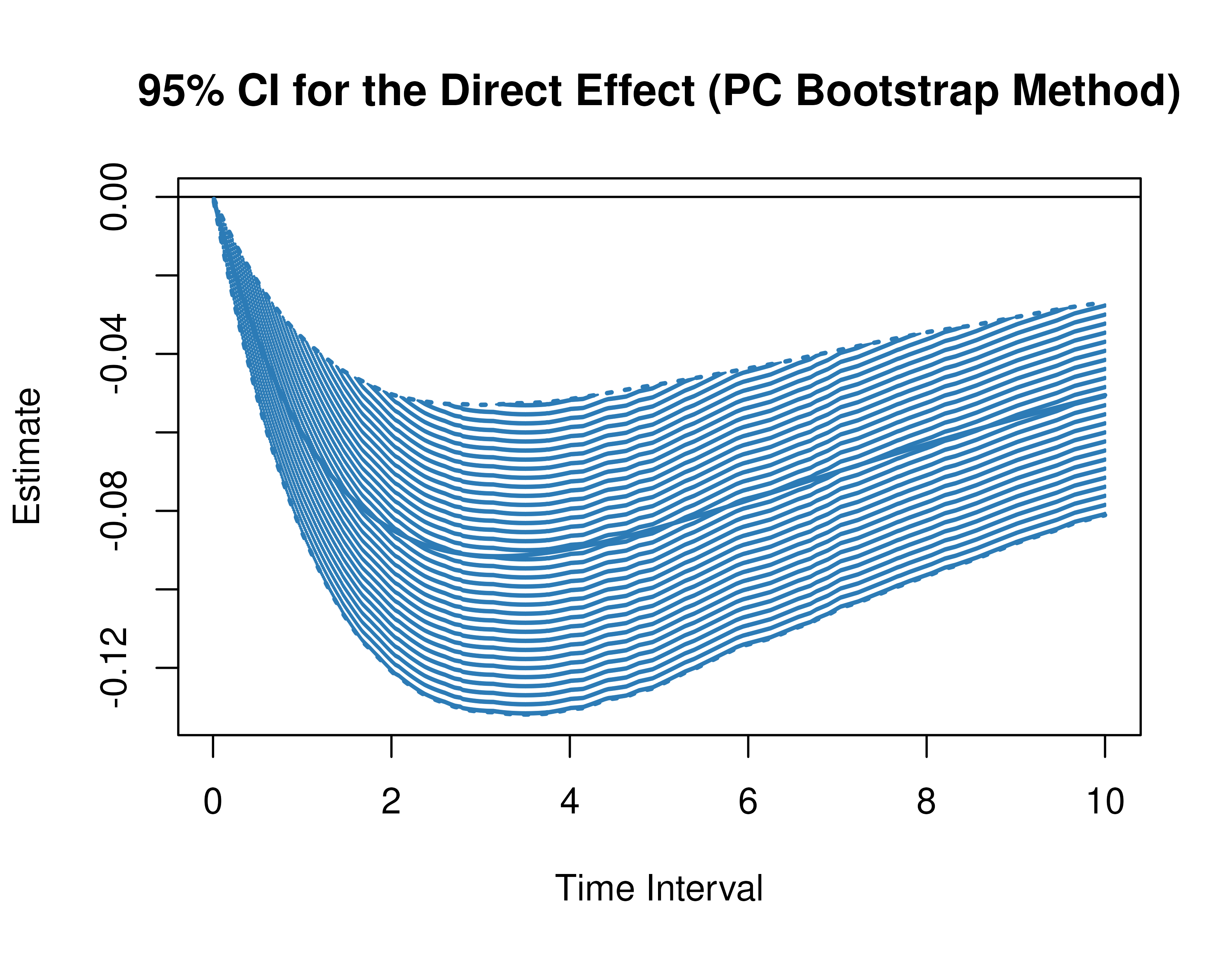
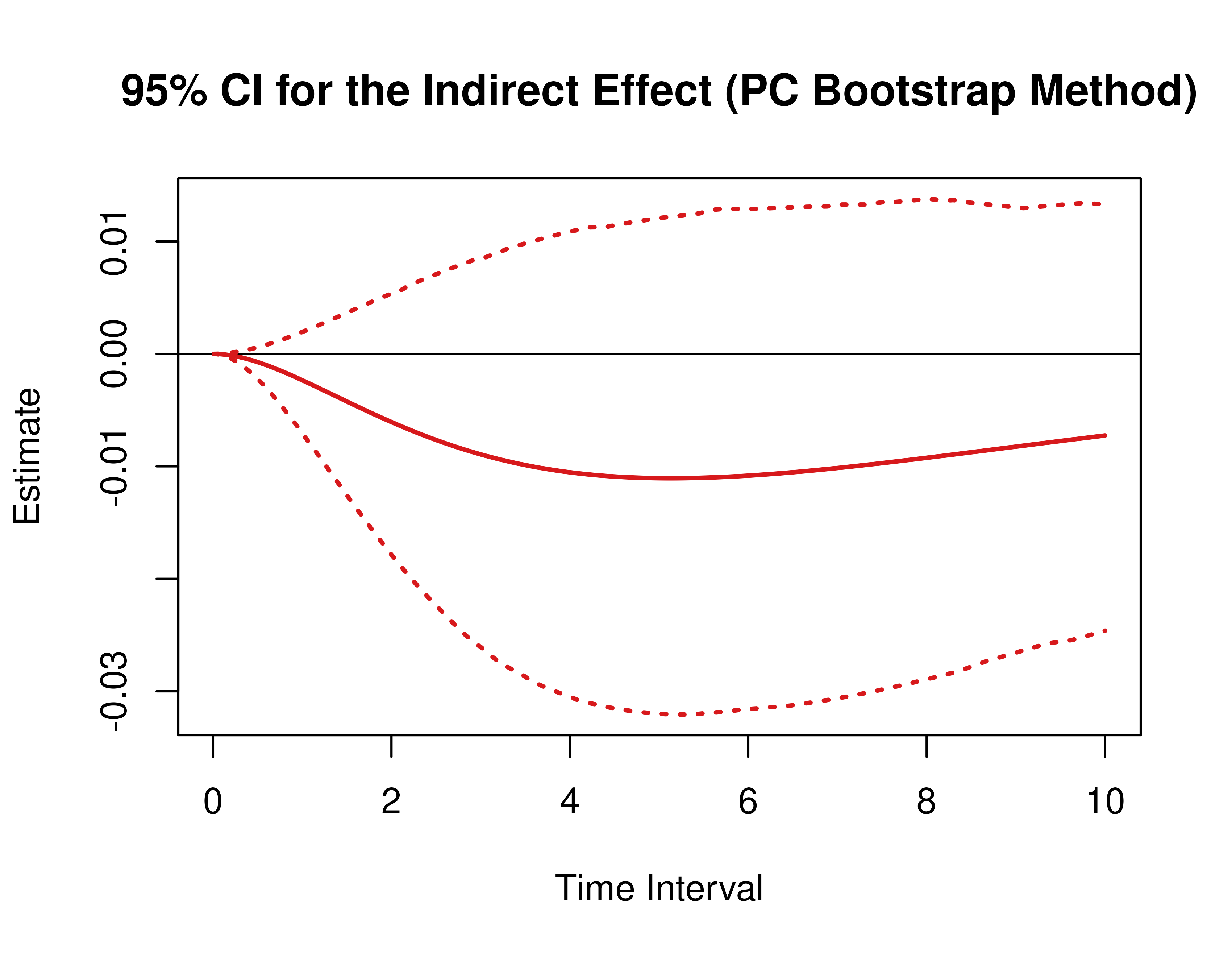
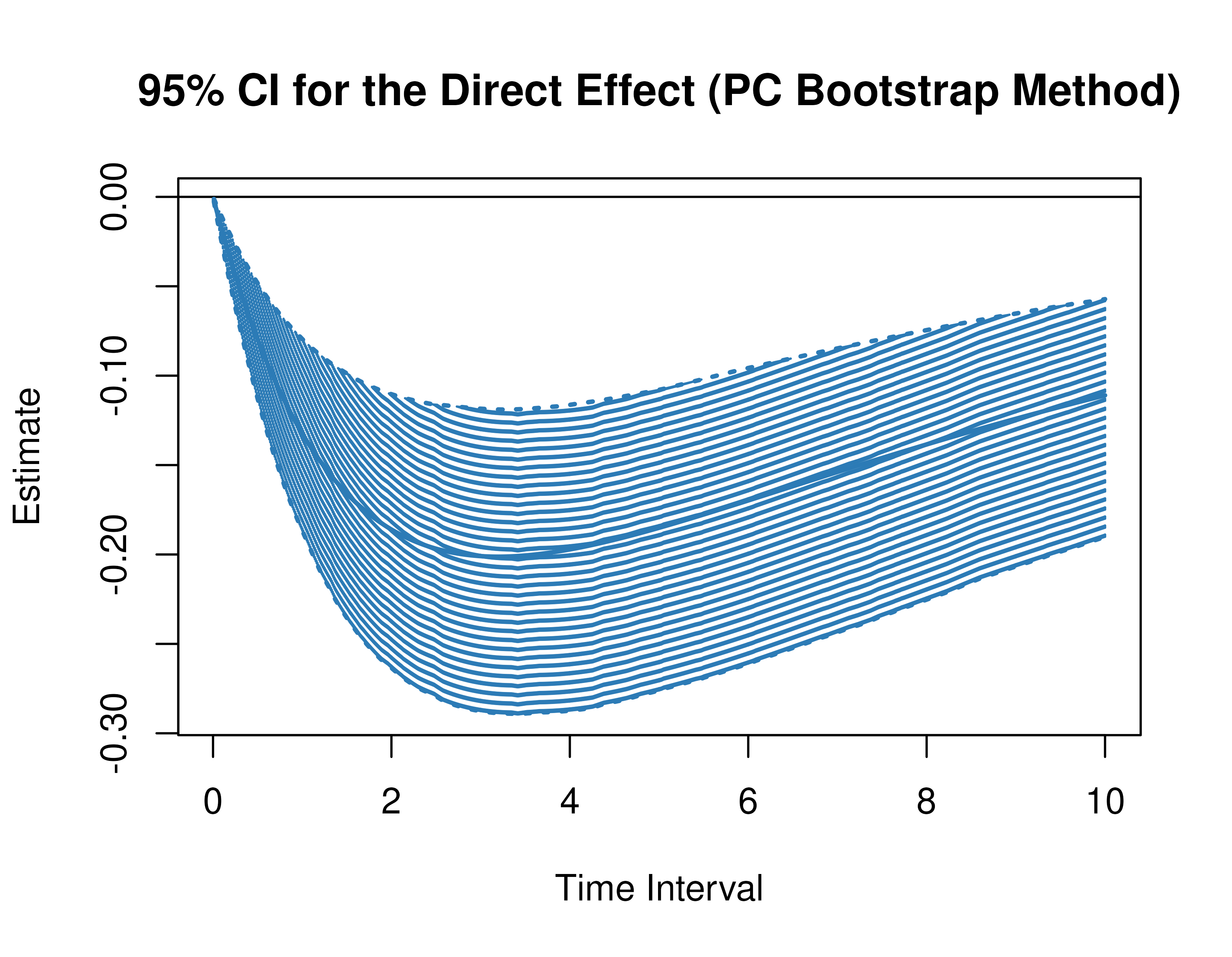
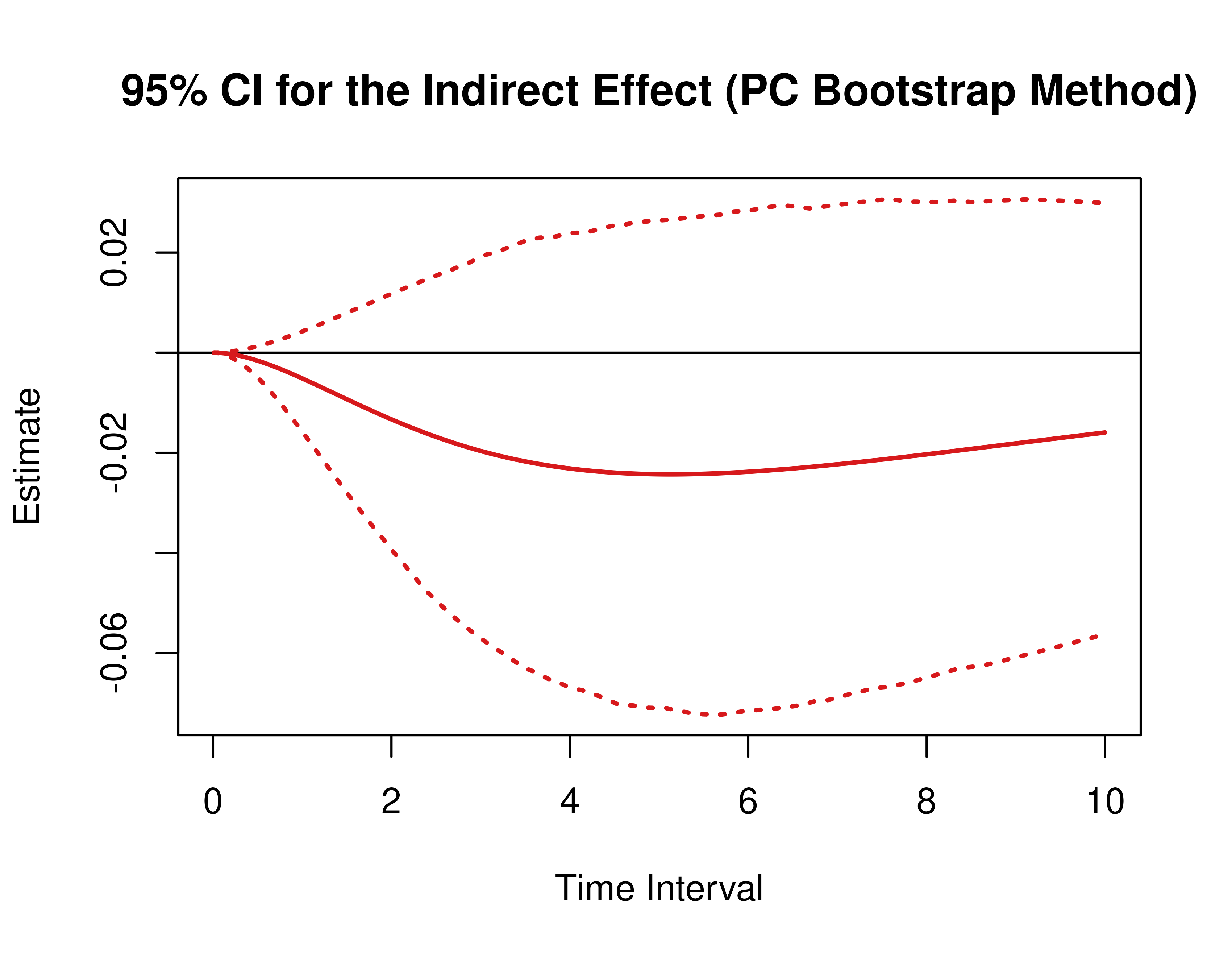
)Using the BootMed function from the cTMed
package, confidence intervals for the direct, indirect, and total
effects for a long sequence of time interval values are generated. This
makes regions of significance more visible. Consider using the
ncores argument to use multiple cores when the
delta_t vector is long. The plot method for
the BootMed presents the regions of significance visually
represented by shaded areas in the plot.
pb <- BootMed(
phi = extract(object = boot, what = "phi"), # extracts the bootstrap drift matrices
phi_hat = phi_hat,
delta_t = delta_t,
from = "conflict",
to = "competence",
med = "knowledge",
ncores = parallel::detectCores()
)
plot(pb)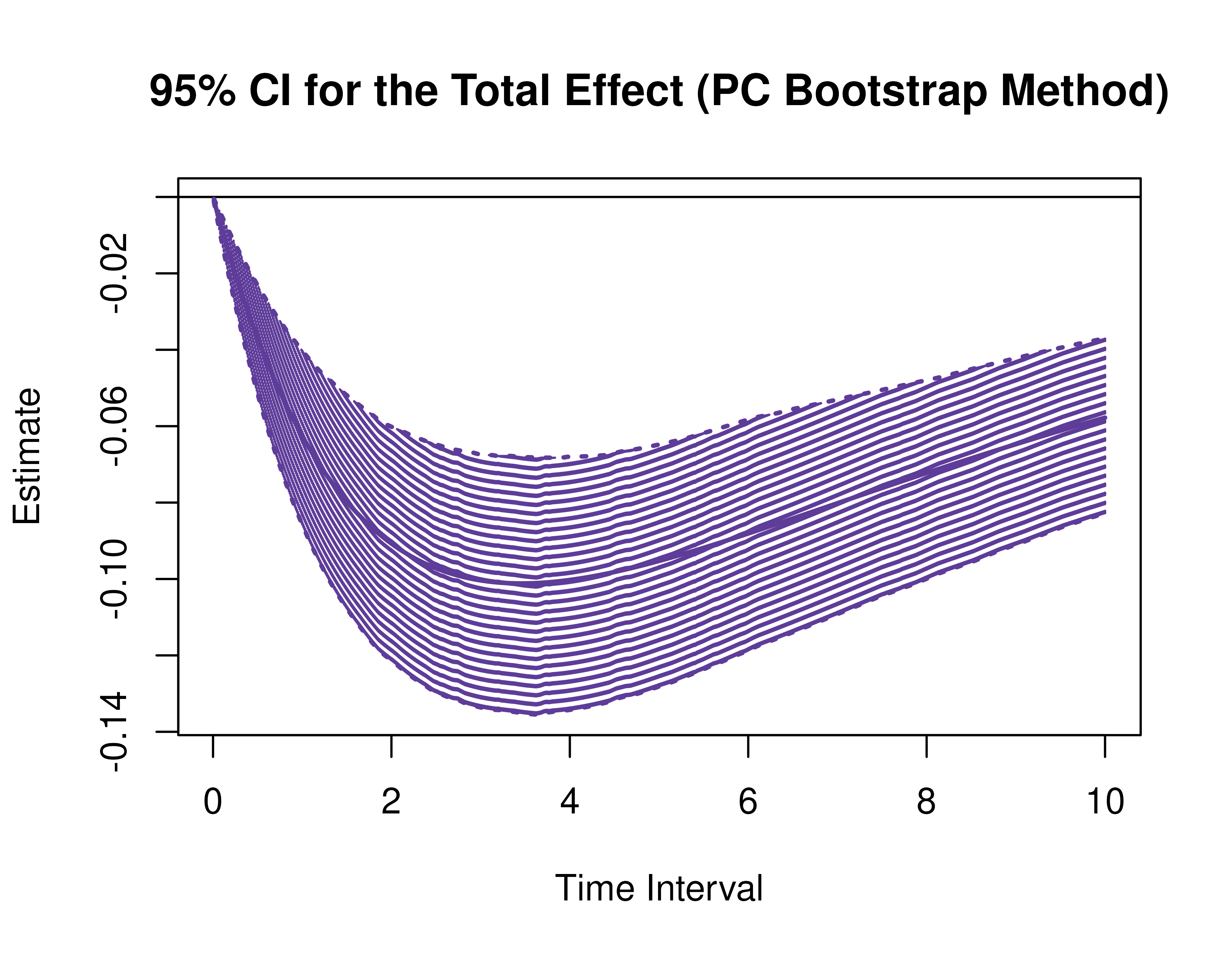
plot(pb, type = "bc") # bias-corrected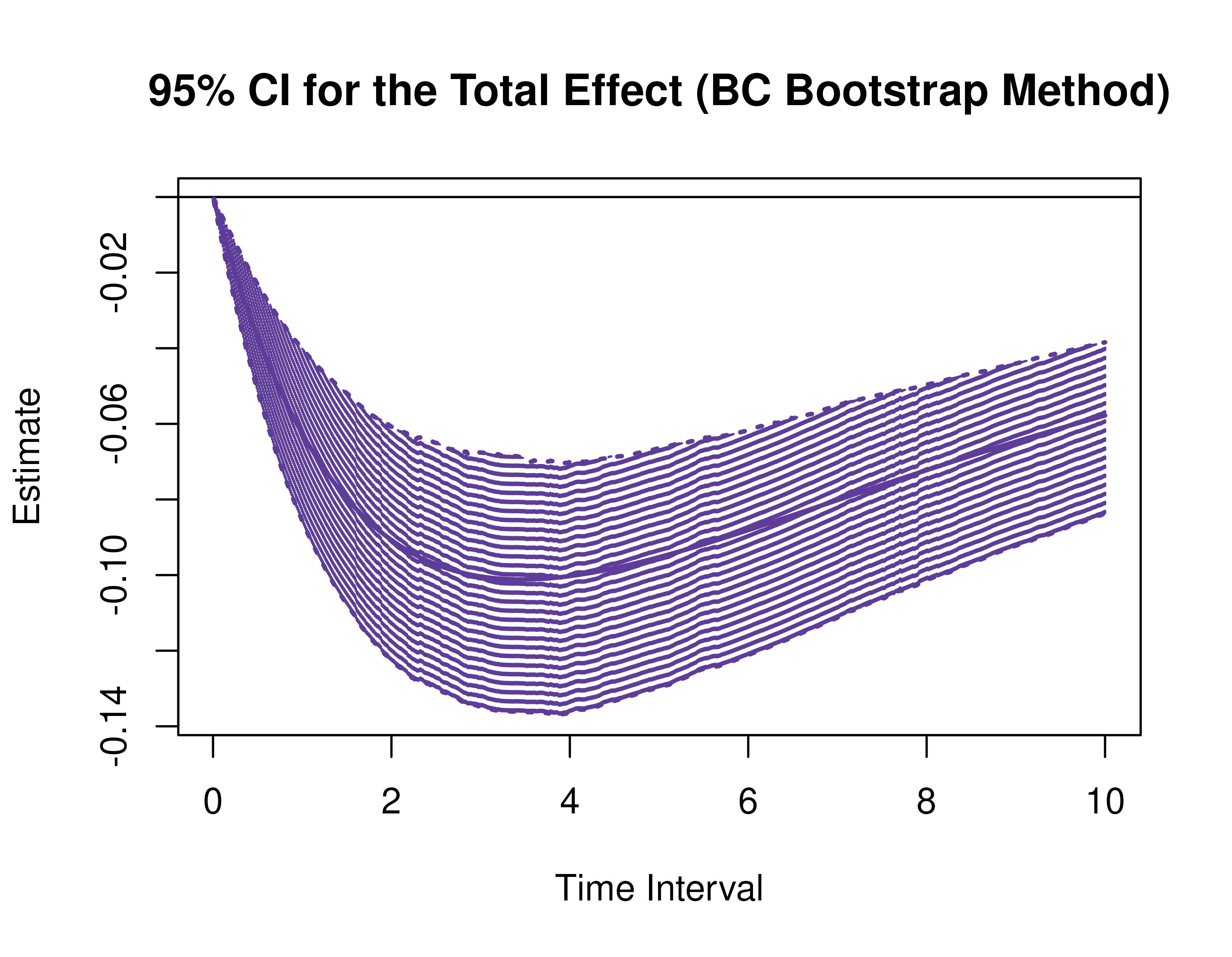
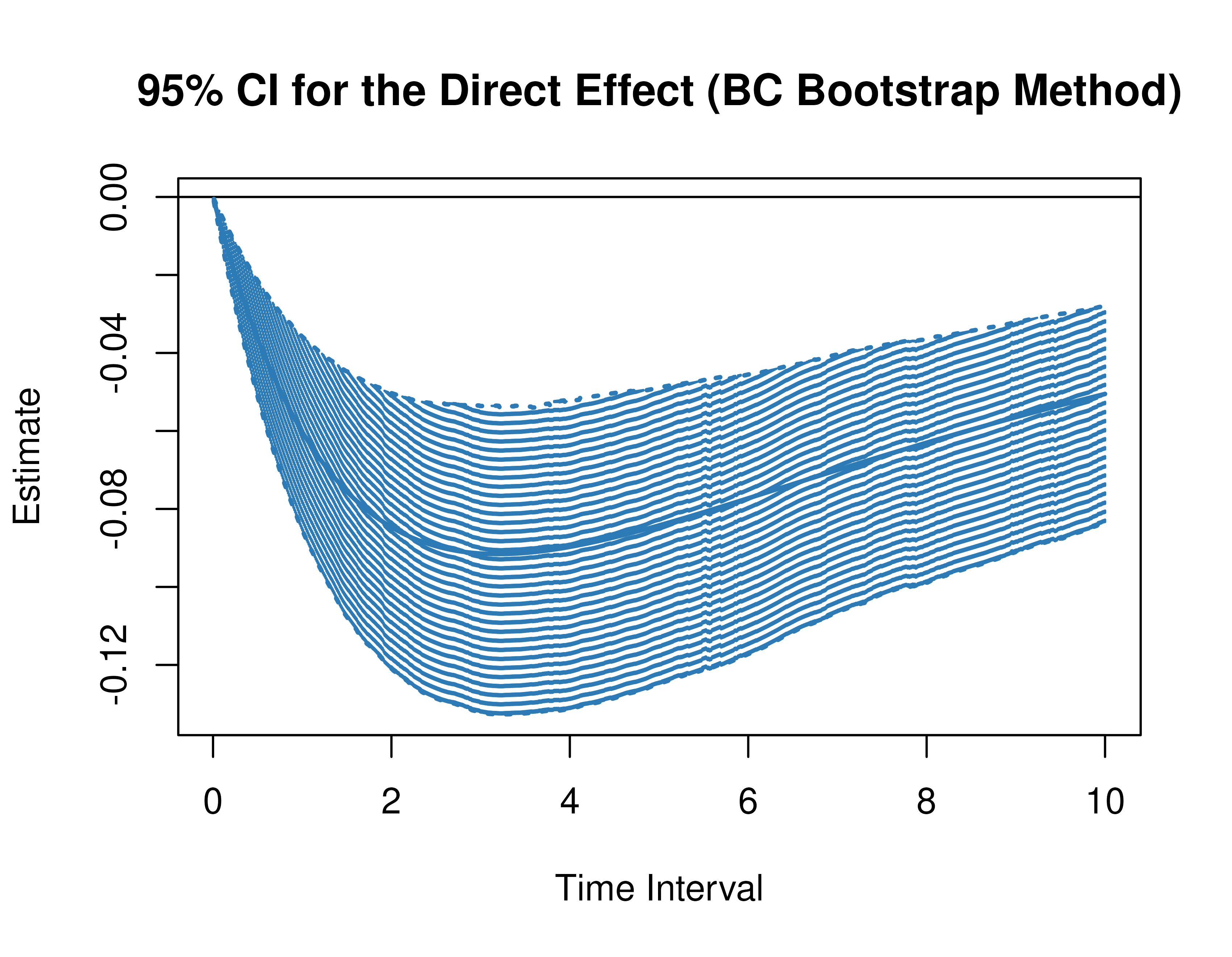
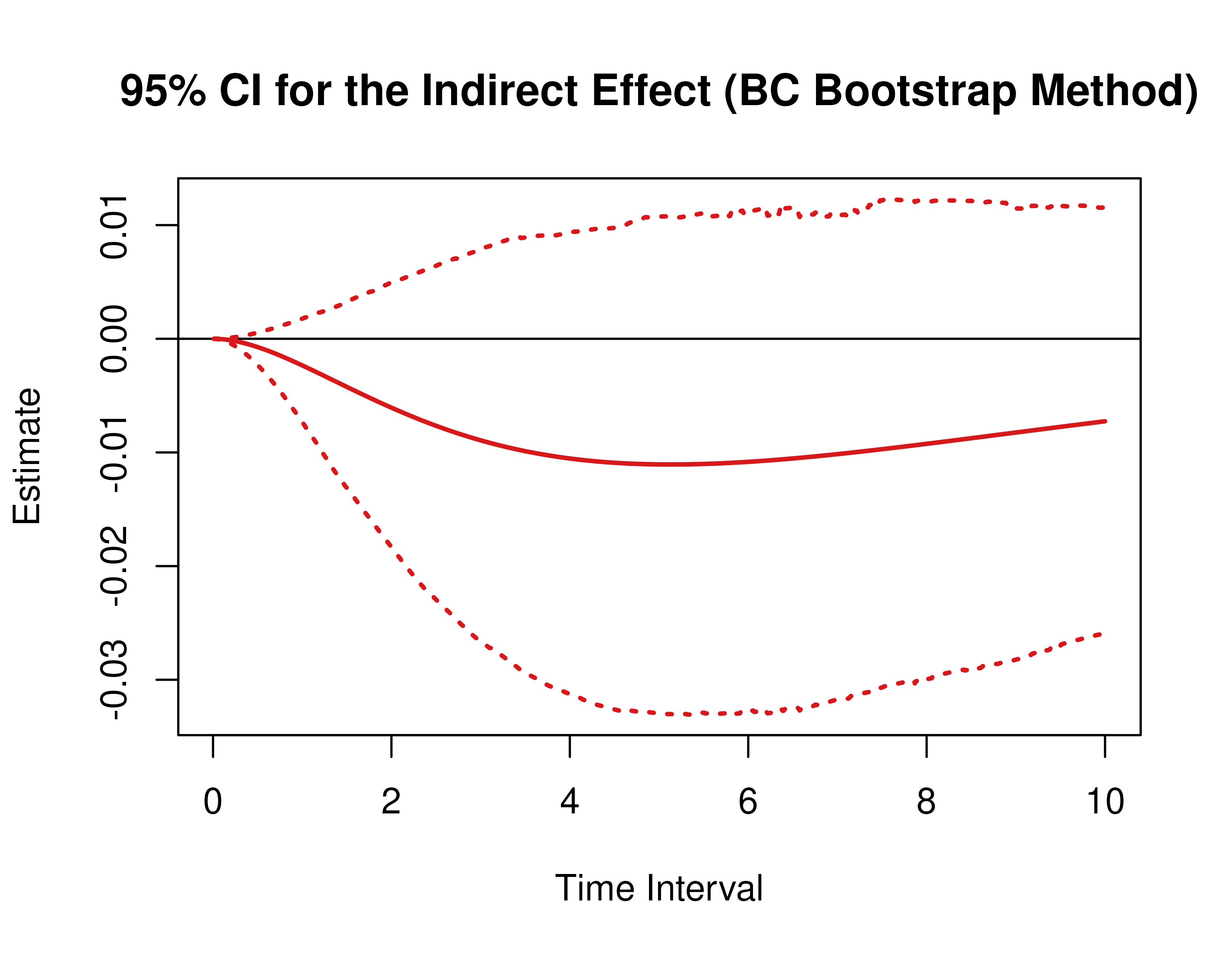
Delta Method Confidence Intervals For The Standardized Direct, Indirect, and Total Effects
Using the DeltaMedStd function from the
cTMed package, confidence intervals for the direct,
indirect, and total effects for a long sequence of time interval values
are generated.
delta_std <- DeltaMedStd(
phi = phi,
sigma = sigma,
vcov_theta = vcov_theta,
delta_t = delta_t,
from = "conflict",
to = "competence",
med = "knowledge",
ncores = parallel::detectCores()
)
plot(delta_std)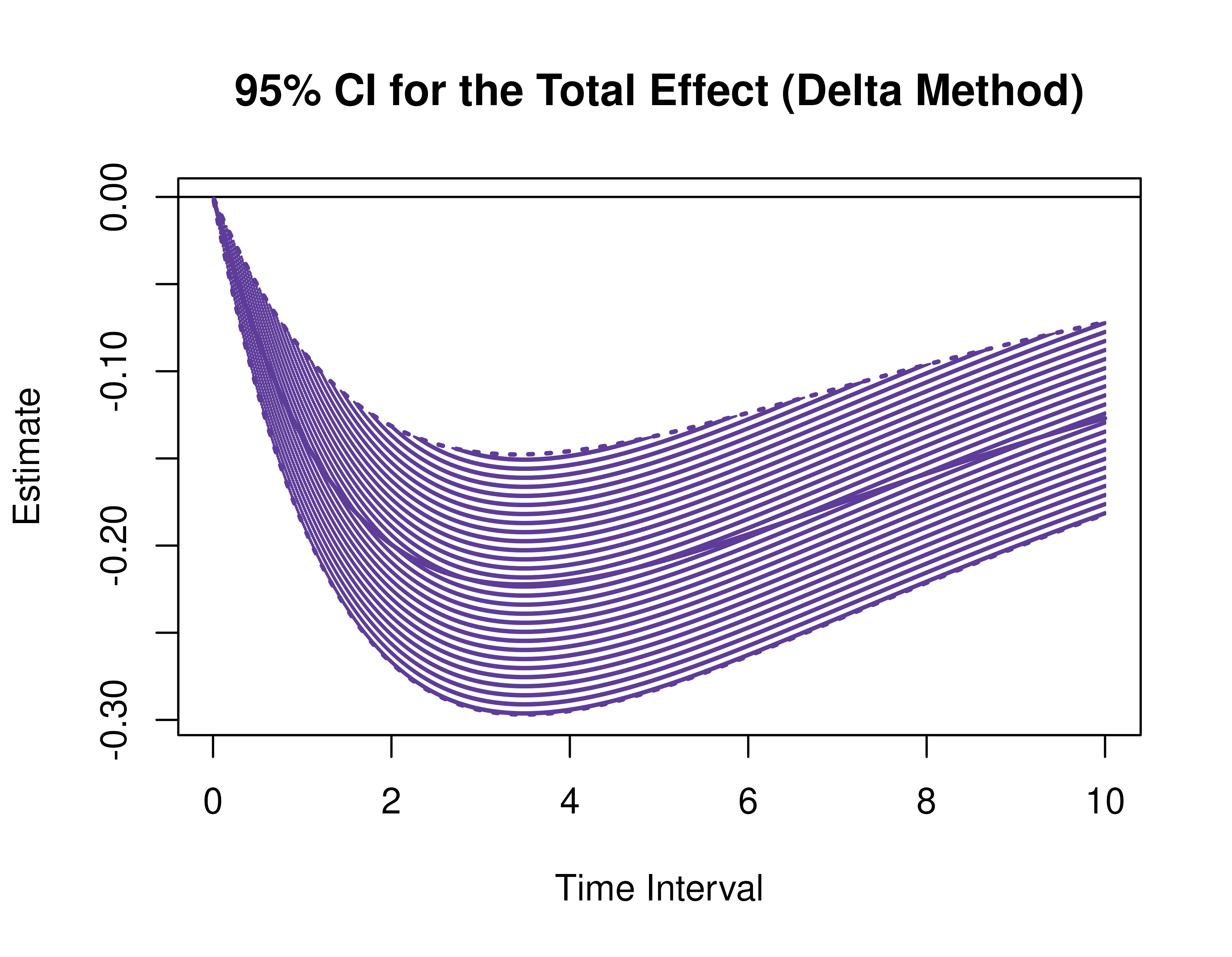
Monte Carlo Method Confidence Intervals For The Standardized Direct, Indirect, and Total Effects
Using the MCMedStd function from the cTMed
package, confidence intervals for the standardized direct, indirect, and
total effects for a time interval of one, two, and three are given
below.
mc_std <- MCMedStd(
phi = phi,
sigma = sigma,
vcov_theta = vcov_theta,
delta_t = delta_t,
from = "conflict",
to = "competence",
med = "knowledge",
R = 20000L,
ncores = parallel::detectCores(),
seed = 42
)
plot(mc_std)
Parametric Bootstrap Method Confidence Intervals For The Standardized Direct, Indirect, and Total Effects
Using the BootMedStd function from the
cTMed package, confidence intervals for the standardized
direct, indirect, and total effects for a time interval of one, two, and
three are given below.
pb <- BootMedStd(
phi = extract(object = boot, what = "phi"), # extracts the bootstrap drift matrices
sigma = extract(object = boot, what = "sigma"), # extracts the bootstrap process noise covariance matrices
phi_hat = phi_hat,
sigma_hat = sigma_hat,
delta_t = delta_t,
from = "conflict",
to = "competence",
med = "knowledge",
ncores = parallel::detectCores()
)
plot(pb)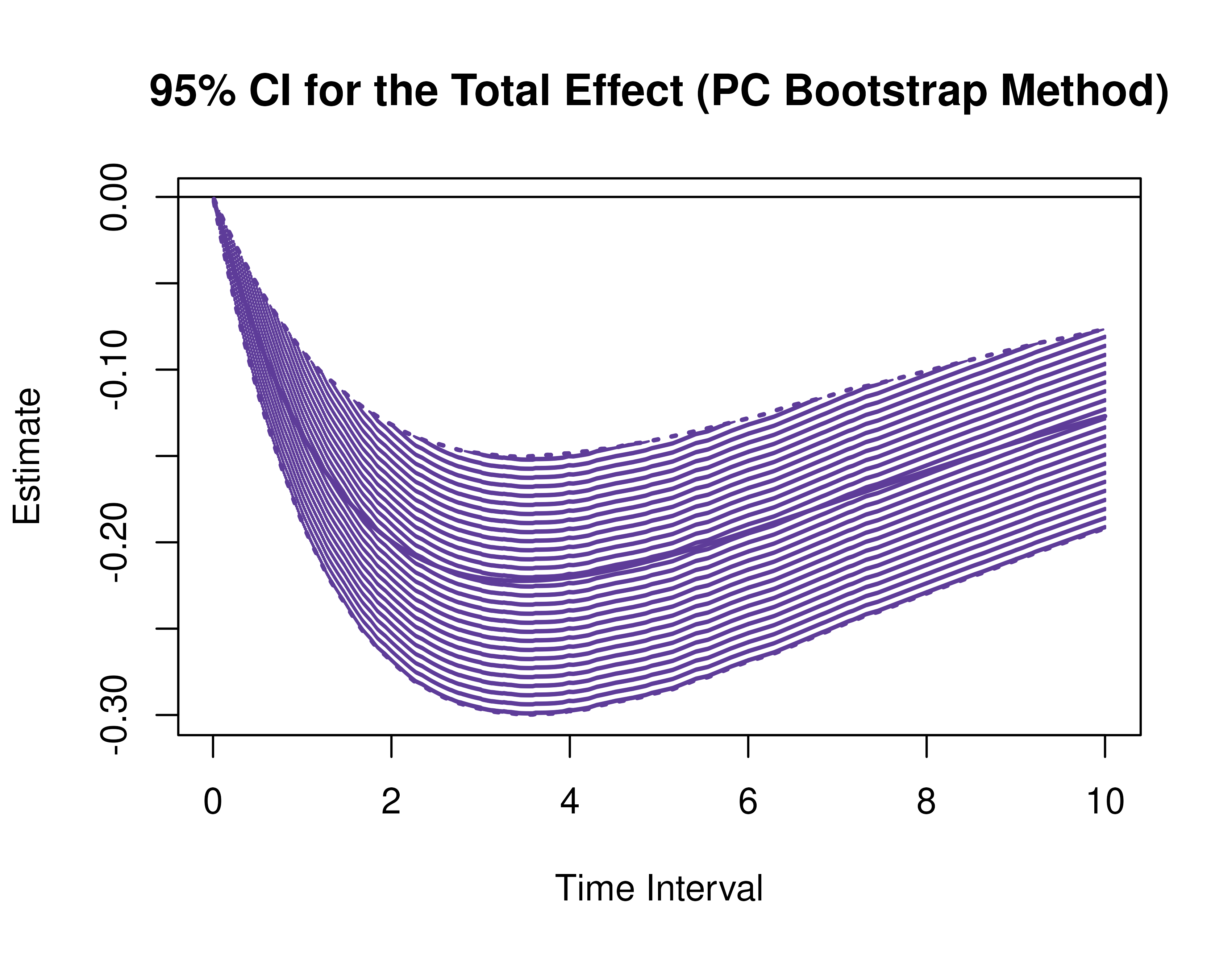
plot(pb, type = "bc")