Illustration 1 - Direct, Indirect, and Total Effects
Ivan Jacob Agaloos Pesigan
Source:vignettes/fig-example-1.Rmd
fig-example-1.RmdThis vignette accompanies Illustration 1. The goal of the
illustration is to calculate the direct, indirect, and total effects
from the continuous-time vector autoregressive model drift matrix
and process noise covariance matrix
for a specific time interval or a range of time intervals. This example
features the Med and MedStd functions from the
cTMed package.
Continuous-Time Vector Autoregressive Model Estimates
The object fit contains the fitted dynr
model. Data is generated using the
manCTMed::IllustrationGenData function. The model was
fitted using the manCTMed::IllustrationFitDynr.
summary(fit)
#> Coefficients:
#> Estimate Std. Error t value ci.lower ci.upper Pr(>|t|)
#> phi_11 -0.1157754 0.0174197 -6.646 -0.1499173 -0.0816334 <2e-16 ***
#> phi_12 0.0203747 0.0550571 0.370 -0.0875352 0.1282846 0.3557
#> phi_13 -0.0070992 0.0535302 -0.133 -0.1120165 0.0978181 0.4472
#> phi_21 -0.1226909 0.0187426 -6.546 -0.1594258 -0.0859560 <2e-16 ***
#> phi_22 -0.8819998 0.0653717 -13.492 -1.0101259 -0.7538737 <2e-16 ***
#> phi_23 0.4737224 0.0594135 7.973 0.3572740 0.5901708 <2e-16 ***
#> phi_31 -0.0877980 0.0178492 -4.919 -0.1227817 -0.0528143 <2e-16 ***
#> phi_32 0.0595814 0.0586861 1.015 -0.0554414 0.1746041 0.1550
#> phi_33 -0.6636176 0.0616495 -10.764 -0.7844485 -0.5427867 <2e-16 ***
#> sigma_11 0.0954899 0.0049239 19.393 0.0858393 0.1051405 <2e-16 ***
#> sigma_12 0.0009624 0.0040987 0.235 -0.0070710 0.0089957 0.4072
#> sigma_13 0.0070297 0.0040069 1.754 -0.0008237 0.0148830 0.0397 *
#> sigma_22 0.1027182 0.0078961 13.009 0.0872422 0.1181942 <2e-16 ***
#> sigma_23 0.0009240 0.0049248 0.188 -0.0087285 0.0105764 0.4256
#> sigma_33 0.0991956 0.0074695 13.280 0.0845558 0.1138355 <2e-16 ***
#> theta_11 0.1016133 0.0014901 68.194 0.0986928 0.1045338 <2e-16 ***
#> theta_22 0.0996380 0.0015668 63.592 0.0965671 0.1027089 <2e-16 ***
#> theta_33 0.0992561 0.0015460 64.202 0.0962260 0.1022862 <2e-16 ***
#> mu0_1 -0.1217526 0.0521444 -2.335 -0.2239538 -0.0195514 0.0098 **
#> mu0_2 0.0160785 0.0279075 0.576 -0.0386192 0.0707762 0.2823
#> mu0_3 0.0204163 0.0288928 0.707 -0.0362125 0.0770451 0.2399
#> sigma0_11 0.3346315 0.0446548 7.494 0.2471097 0.4221533 <2e-16 ***
#> sigma0_12 -0.0644525 0.0177503 -3.631 -0.0992424 -0.0296626 0.0001 ***
#> sigma0_13 -0.0519690 0.0179974 -2.888 -0.0872433 -0.0166946 0.0019 **
#> sigma0_22 0.0691741 0.0127707 5.417 0.0441439 0.0942043 <2e-16 ***
#> sigma0_23 0.0364469 0.0098132 3.714 0.0172134 0.0556805 0.0001 ***
#> sigma0_33 0.0792446 0.0137057 5.782 0.0523819 0.1061072 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> -2 log-likelihood value at convergence = 32662.46
#> AIC = 32716.46
#> BIC = 32919.11Extract Elements of the Drift Matrix and the Process Noise Covariance Matrix
We extract the elements of the drift matrix and the process noise
covariance matrix from the fit object.
# drift matrix
phi <- matrix(
data = coef(fit)[
c(
"phi_11",
"phi_21",
"phi_31",
"phi_12",
"phi_22",
"phi_32",
"phi_13",
"phi_23",
"phi_33"
)
],
nrow = 3
)
# column names and row names are needed to define the indirect effects
colnames(phi) <- rownames(phi) <- c(
"conflict",
"knowledge",
"competence"
)Direct, Indirect, and Total Effects of a Time Interval of Three
Using the Med function from the cTMed
package, the direct, indirect, and total effects for a time interval of
three are given below.
Med(
phi = phi,
from = "conflict",
to = "competence",
med = "knowledge",
delta_t = 3
)
#>
#> Total, Direct, and Indirect Effects
#>
#> interval total direct indirect
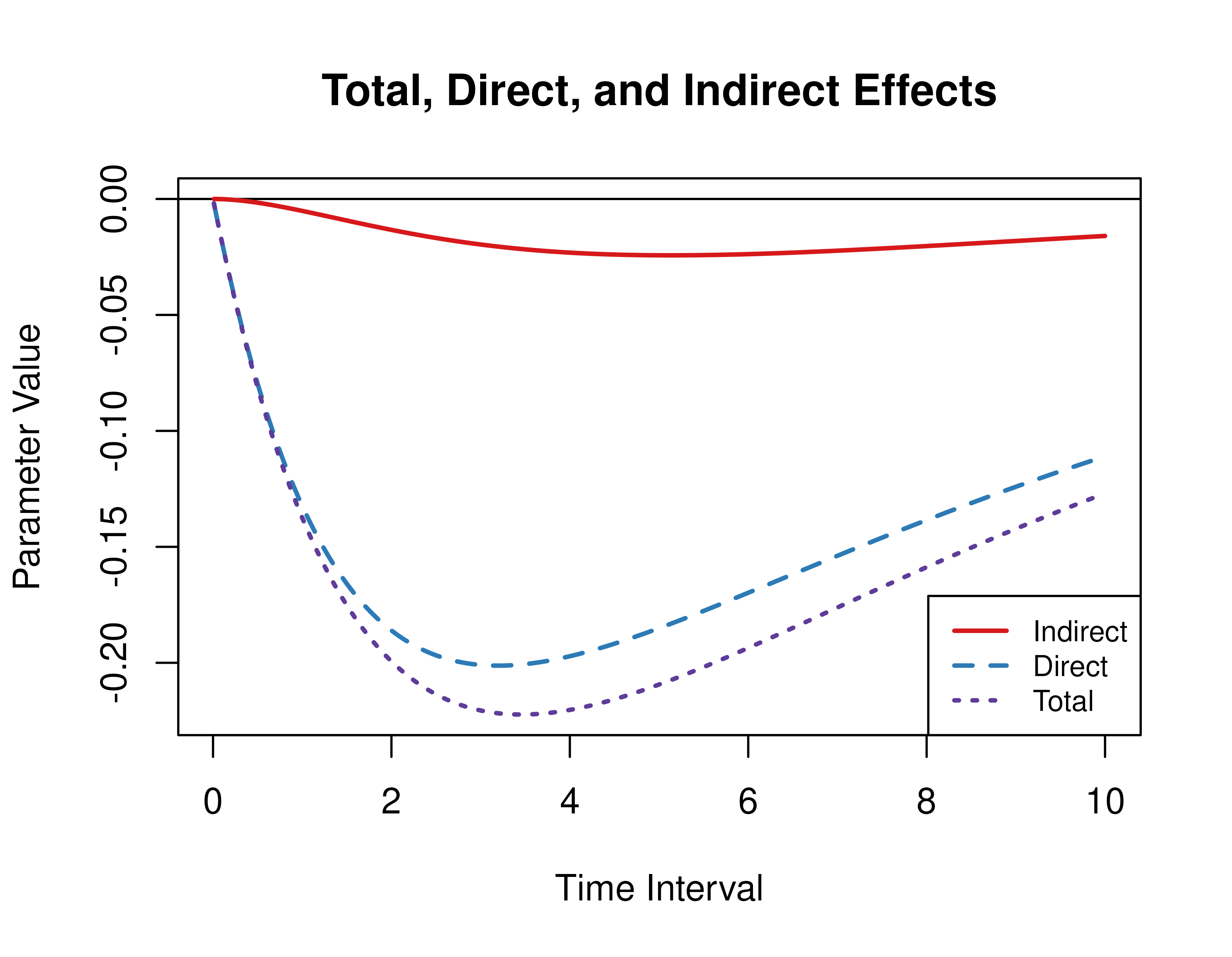
#> [1,] 3 -0.1004 -0.0914 -0.0089Direct, Indirect, and Total Effects of a Time Interval of Zero to Ten
Using the Med function from the cTMed
package, the direct, indirect, and total effects for a range of time
interval values from 0 to 10 are plotted below.
med <- Med(
phi = phi,
from = "conflict",
to = "competence",
med = "knowledge",
delta_t = seq(from = 0, to = 10, length.out = 1000)
)
plot(med, legend_pos = "bottomright")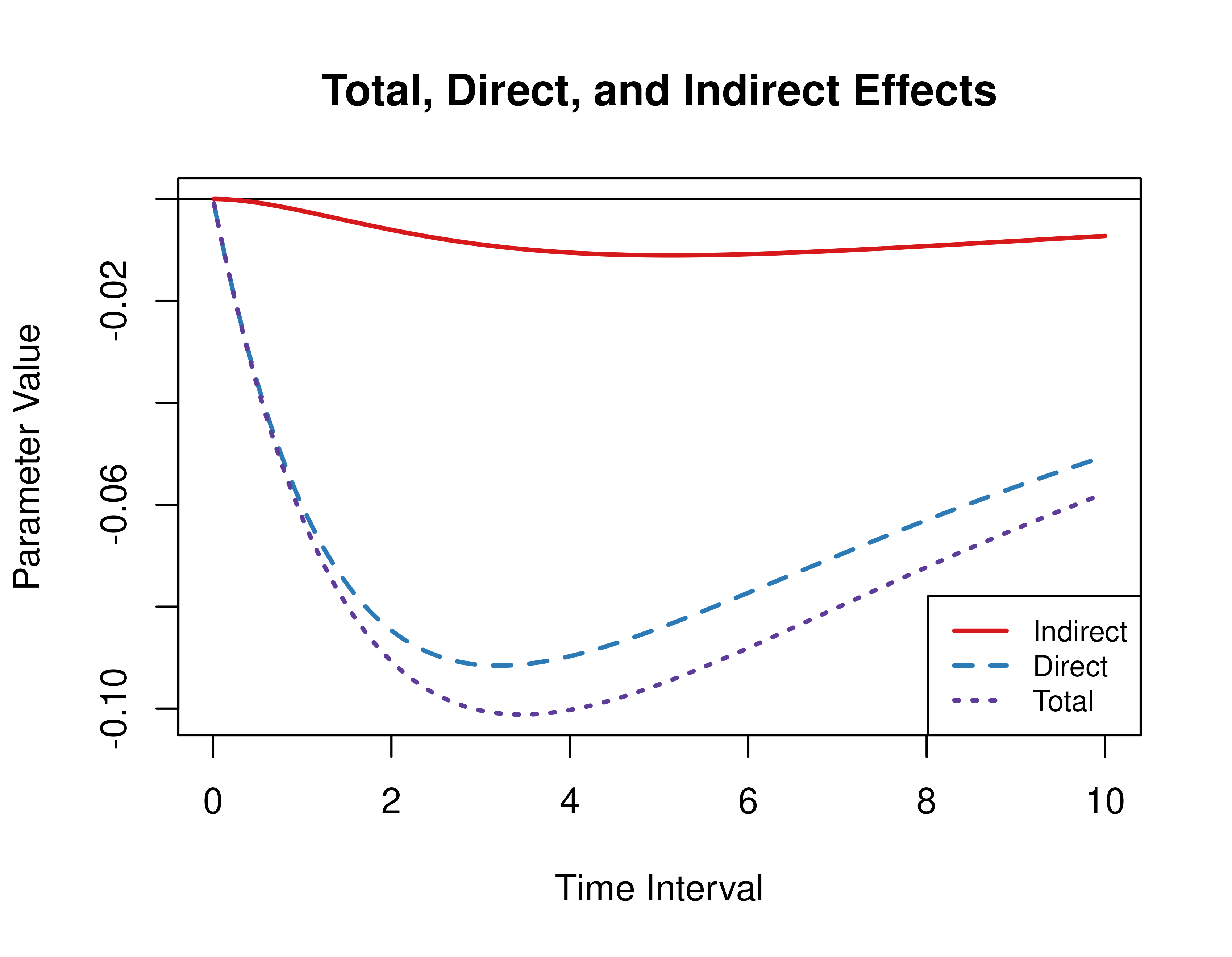
Standardized Direct, Indirect, and Total Effects of a Time Interval of Three
Using the MedStd function from the cTMed
package, the standardized direct, indirect, and total effects for a time
interval of three are given below.
MedStd(
phi = phi,
sigma = sigma,
from = "conflict",
to = "competence",
med = "knowledge",
delta_t = 3
)
#>
#> Total, Direct, and Indirect Effects
#>
#> interval total direct indirect
#> [1,] 3 -0.2206 -0.2009 -0.0196Standardized Direct, Indirect, and Total Effects of a Time Interval of Zero to Ten
Using the Med function from the cTMed
package, the standardized direct, indirect, and total effects for a
range of time interval values from 0 to 10 are plotted below.
med_std <- MedStd(
phi = phi,
sigma = sigma,
from = "conflict",
to = "competence",
med = "knowledge",
delta_t = seq(from = 0, to = 10, length.out = 1000)
)
plot(med_std, legend_pos = "bottomright")